

Next: Specification of Potentials
Up: OPTIM3 User Guide
Previous: The odata file
Keywords
The following keywords are recognised, where n, x and exec are integer,
real and character data, respectively.
- 2D: restrict the system to the
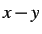 plane.
plane.
- ACE n: for use with CHARMM runs that employ the ACE implicit solvent model.
n specifies the number of calls to the CHARMM energy and gradient for which lists
are held fixed. The default value of n is 50.
Note: ENDNUMHESS must be used with the ACE solvent model.
Otherwise OPTIM will attempt to use incorrect second derivatives.
ENDNUMHESS will be needed in the odata.start, odata.finish
and odata.connect files.
For odata.start and odata.finish the keyword ENDHESS
must also be included.
- ADM n: will cause the
interatomic distance matrix to be printed every n cycles;
the default for n is 20. This matrix is not printed by default.
- ALPHA: sets exponent value for averaged Gaussian and Morse potentials,
default value is 6.
- ALLPOINTS: turns on printing of coordinates to file points for
all intermediate configurations. Default is false.
- AMBER: specifies a calculation with the AMBER potential. This requires
auxiliary files amber.dat and coords.amber in the same directory.
- ANGLEAXIS: specifies angle/axis coordinates for rigid body TIP potentials.
- ARCTOL tol: specifies the accuracy tolerance for computing the
inverse arc length used in the cubic spline interpolated string methods for
double-ended transition state searches. The default is
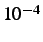 .
.
- AXIS n: specifies the highest symmetry axis to search for in
routine symmetry; default is six.
- BFGSCONV gmax:
gmax is the convergence criterion
for the root-mean-square gradient, default
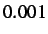 .
This is also the convergence criterion
for the subspace minimisations in hybrid EF/BFGS transition state searches, for use with BFGSTS.
.
This is also the convergence criterion
for the subspace minimisations in hybrid EF/BFGS transition state searches, for use with BFGSTS.
- BFGSMIN gmax: instructs the program to perform an LBFGS minimisation.
gmax is the convergence criterion
for the root-mean-square gradient, default
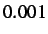 .
.
- BFGSSTEP: instructs the program to step off a saddle point along the
eigenvector corresponding to the smallest negative eigenvalue without
diagonalizing the Hessian to find this eigenvector. It is possible to step off
parallel and antiparallel to this eigenvector by specifying positive or negative values
for the MODE parameter. BFGSTS need not be set, but see also
PATH and CONNECT. After taking the first step the program switches to
energy minimisation using whichever algorithm is specified, e.g. BFGSMIN,
SEARCH 0, SEARCH 6, RKMIN, BSMIN, etc.
- BFGSSTEPS n: sets the maximum number of steps allowed in LBFGS minimisations.
Default value is the value of the STEPS parameter.
However, for NEWNEB this parameter is specified via the NEWCONNECT or
NEWNEB line.
- BFGSTS nevs ncgmax1 ncgmax2 ceig nevl: instructs the program to perform a
hybrid BFGS/eigenvector-following transition state search.
nevs is an integer that defines the largest number of iterations allowed in the
calculation of the smallest Hessian eigenvalue; default 500.
ncgmax1 is an integer that defines the largest number of BFGS steps
allowed in the subspace minimisation before the eigenvalue is deemed to have converged; default 10.
ncgmax2 is an integer that defines the largest number of BFGS steps
allowed in the subspace minimisation after the eigenvalue is deemed to have converged; default 100.
This parameter is ignored if NOIT is set.
The eigenvalue is deemed to be converged if the modulus of the overlap between the corresponding
eigenvector and that saved from the previous step exceeds 0.999 and we have the right number of
negative eigenvalues.
ceig is a double precision parameter that sets the convergence criterion for
the calculation of the smallest non-zero Hessian eigenvalue.
If NOHESS is set ceig is compared to the RMS `force' corresponding to the
Rayleigh-Ritz expectation value that is minimised to get the smallest eigenvalue.
If the Hessian is used via iteration or NOIT
then ceig is compared to the percentage change in the eigenvalue between successive
steps. The default value is 1.0, which is more appropriate to the latter case. Smaller
values are probably necessary in BFGS/BFGS calculations.
nevl is an integer that defines the largest number of iterations allowed in the
calculation of the largest Hessian eigenvalue; default 100. Not needed if NOHESS
of NOIT is set.
- BINARY ntypea epsab epsbb sigmaab sigmabb: specifies a binary Lennard-Jones
system for use with the LP or LS atom types. ntypea is the number of type
A atoms--the rest are assumed to be type B and appear at the end of the list
of coordinates.
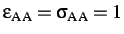 define the units of energy and length,
and epsab=
define the units of energy and length,
and epsab=
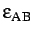 , epsbb=
, epsbb=
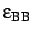 ,
sigmaab=
,
sigmaab=
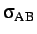 , sigmabb=
, sigmabb=
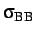 .
.
- BLN rkr rkt: general BLN protein bead model.[18,19]
rkr and rkt are the spring constants for bond length and bond angle terms.
An auxiliary file BLNsequence is required to specify all the other parameters and
the sequence, along with a code to define the secondary structure template that changes
some of the parameters.
For example, the format of BLNsequence for protein L is:
comment: LJREP_BLN >0 and LJATT_BLN <0 for B-B, L-L and L-B, N-L and N-B and N-N
1.0D0 -1.0D0
0.33333333333333D0 0.33333333333333D0
1.0D0 0.0D0
comment: coefficients A, B, C, D of
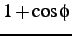 ,
, 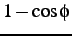 ,
, 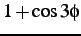 and
and
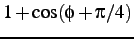 comment: for Helical, Extended and Turn residues in order, four per line
0.0D0 1.2D0 1.2D0 1.2D0
0.9D0 0.0D0 1.2D0 0.0D0
0.0D0 0.0D0 0.2D0 0.0D0
LBLBLBLBBNNNBBBLBLBBBNNNLLBLLBBLLBNBLBLBLBLNNNLBBLBLBBBL
EEEEEETEHTHEEEEEEEEHHEHHHHHHHHHHEHTEEEEEEETTTEEEEEEEE
comment: for Helical, Extended and Turn residues in order, four per line
0.0D0 1.2D0 1.2D0 1.2D0
0.9D0 0.0D0 1.2D0 0.0D0
0.0D0 0.0D0 0.2D0 0.0D0
LBLBLBLBBNNNBBBLBLBBBNNNLLBLLBBLLBNBLBLBLBLNNNLBBLBLBBBL
EEEEEETEHTHEEEEEEEEHHEHHHHHHHHHHEHTEEEEEEETTTEEEEEEEE
- BSMIN gmax eps: calculates a steepest-descent path using gradient only
information with convergence criterion gmax for the RMS force and initial
precision eps. The Bulirsch-Stoer algorithm is used.
- BULK: specifies that a periodically repeated supercell is being used.
- CADPAC system exec: tells the program to read derivative information in
CADPAC format. system is a string to identify the system and exec is
the name of an executable that will generate a CADPAC input deck from a points file.
If exec is omitted its name is assumed to be editit.system.
- CALCDIHE: calculates an order parameter for CHARMM and UNRES with respect to
a reference structure in file ref.crd.
- CALCRATES temp h: rate constants will be calculated for each transition state
found in a CONNECT or PATH run, or using saved information in an
old path.info file if READPATH is specified. Both forward and backward canonical
rate constants are calculated from transition state theory at temperature temp (default
temp=1), and values are given
for both quantum and classical harmonic vibrational partition functions. h is the
value of Planck's constant in suitable reduced units for the corresponding potential (default value 1).
- CAPSID rho eps r h: specifies a potential for rigid pentagonal pyramids with
Morse parameter rho, repulsive strength eps, radius r, and height h (in
units of r).
h may be omitted, in which case it defaults to 0.5.
- CASTEP system: tells the program to use CASTEP to calculate energies and
forces with input files based on the system name system. Periodic boundary
conditions are assumed, so projection is not performed to remove overall rotation.
- CASTEPC system: tells the program to use CASTEP to calculate energies and
forces with input files based on the system name system.
In this case the system is assumed to be a cluster, so that both translational and rotational
degrees of freedom correspond to zero Hessian eigenvalues at a stationary point.
- CHARMM: specifies that the CHARMM potential is used. An auxiliary file
input.crd is required. CHARMM must be the last OPTIM directive in the
odata file. The remaining content of odata consists of CHARMM keywords and
setup information.
- CHARMMTYPE n: n specifies the particular CHARMM potential.
n=1, 2 and 3 correspond to CHARMM22 with ACE-type capping groups, CHARMM19 with ACE-type
capping groups, and CHARMM19 with no capping groups, respectively.
n also determines which atoms are uses to set the
seed atoms for rebuilding after internal coordinate manipulation.
- CHECKINDEX nevs ceig nevl: instructs the program to check the Hessian
index after a BFGS minimisation or BFGS hybrid transition
state search. This keyword also works with NOHESS.
The parameters are the same as for BFGSTS below, and need not be set
if they have already been specified by that keyword.
- CHECKCHIRALITY: checks that all residues are in the L-isomer
state and rejects transition states and minima that are not.
- CHECKCONT: if CHECKINDEX is specified then CHECKCONT
instructs the program to take a pushoff and continue if convergence to a stationary
point with the wrong Hessian index is detected.
- COLDFUSION thresh: if the energy falls below threshold thresh then
cold fusion is assumed to have occurred and geometry optimisation stops.
- COMMENT or NOTE: the rest of the line is ignored.
- CONNECT n: find a min-sad-min-
 -min sequence connecting
the initial minimum in odata
(or auxiliary files for CHARMM, UNRES, etc.) to the final minimum in finish,
giving up if more than n transition states are needed (default n=100). CONNECT
generally needs to be augmented by other keywords to specify how the transition
state geometries should be guessed (NEB, NEWNEB, FIXD
or GUESSTS), how the transition
state searches should be performed (SEARCH 2 or BFGSTS, with or
without NOHESS, NOIT etc.), and how that pathways should be
calculated (SEARCH 0, 6 or 7, BFGSMIN, RKMIN or BSMIN).
A summary file will be produced if DUMPPATH or DUMPALLPATHS is specified. An xyz file
for the overall path will be printed to path.xyz and the energy as a
function of the path length is printed to EofS. The corresponding xyz and energy
files for the individual steps in the path are numbered path.
-min sequence connecting
the initial minimum in odata
(or auxiliary files for CHARMM, UNRES, etc.) to the final minimum in finish,
giving up if more than n transition states are needed (default n=100). CONNECT
generally needs to be augmented by other keywords to specify how the transition
state geometries should be guessed (NEB, NEWNEB, FIXD
or GUESSTS), how the transition
state searches should be performed (SEARCH 2 or BFGSTS, with or
without NOHESS, NOIT etc.), and how that pathways should be
calculated (SEARCH 0, 6 or 7, BFGSMIN, RKMIN or BSMIN).
A summary file will be produced if DUMPPATH or DUMPALLPATHS is specified. An xyz file
for the overall path will be printed to path.xyz and the energy as a
function of the path length is printed to EofS. The corresponding xyz and energy
files for the individual steps in the path are numbered path. .xyz and EofS.
.xyz and EofS.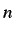 for
transition state
for
transition state 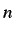 .
.
- CONSEC nstart nend nstart nend etc.: construct linearly interpolated, internal coordinate
transition state guesses and apply the stopping criterion in a CONNECT run over a chosen
segment of protein specified by pairs of residue numbers
(nstart,nend). Up to ten distinct sections can be chosen.
- CONVERGE x y NOINDEX: change the default convergence conditions
for eigenvector-following and second order steepest-descent calculations
described in §5. If one number is supplied then convergence depends only upon
the maximum unscaled step falling below the specified value (and the right number
of negative Hessian eigenvalues). If two numbers are supplied the second is the
required threshold for the RMS force which must also be satisfied.
If the string NOINDEX is present then the Hessian index isn't checked.
- COPYRIGHT: prints copyright information.
- CPMD system: tells the program to use CPMD to calculate energies and
forces with input file system; bulk boundary conditions.
- CPMDC system: tells the program to use CPMD to calculate energies and
forces with input file system; cluster boundary conditions.
- CUBIC: if the three box lengths are equal to start with in a PV run,
and the keyword CUBIC is included, then a cubic box is maintained.
- CUBSPL: For the growing string or evolving string double-ended
transition state search methods, use a cubic spline interpolation between
the image points.
- D5H x: add a decahedral field of strength x to the potential.
- DCHECK ON/OFF: turns on/off warning messages when atoms
approach to within
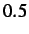 distance units. Default is ON.
distance units. Default is ON.
- DEBUG: causes unscaled steps, gradients, Hessian eigenvalues and
summary information to be printed every cycle for SEARCH 0-7 runs. Turned off by default.
Also produces much more printing for individual steps in CONNECT runs.
- DESMDEBUG: Produces extra printing for the double-ended
transition state search method runs (DNEB, GS or ES).
- DFTB: specifies that the tight-binding code of Walsh and Wales[20]
is used.
- DGUESS dguess1 dguess2 dguess3 dguess4: initial guess for diagonal elements of the inverse
Hessian, used whenever the BFGS optimiser is reset. dguess1 is used in
geometry optimisations, dguess2 is used for minimisation in
eigenvalue calculations, dguess3 is used for DNEB optimisation
steps, and dguess4 is used for string method optimisation steps.
The default values are dguess1=dguess2=0.1, dguess3=dguess4=0.001
- DIJKSTRA [EXP/n] [INTERP x EXP/n]: specifies that the missing connection
Dijkstra-based algorithm[21] should be used to choose minima for connection in cycles
of NEWCONNECT.
The edge weights are based on the smallest distance between minima exponentiated (for EXP) or
raised to the integer power n.
Alternatively, the additional keyword INTERP specifies an edge weight based on
the highest energy difference found by a linear interpolation between the minima at regular spacings
of x.
The final EXP or integer n specifies whether this highest energy is exponentiated
or raised to the power n to obtain the edge weight.
- DOUBLE: adds a double-well potential as per J. Chem. Phys.,
110, 6617, 1999. The
energy and derivatives add to the potential specified by the atom
type. Only atom type
`LS' (not binary) has the 1,2 interaction removed as well.
At low density some atoms may not interact with any others and
problems result due to additional zero eigenvalues.
- DQAGKEY n: n (integer between 1 and 6) indicates the number of local
integration points used for calculating cubic spline arc lengths in the string methods.
Default value is 6, which corresponds to 30-61 points.
- DRAG: obsolete method that transforms one end point into another
by progressively increasing a spring potential that attracts the system to the
final geometry.
- DUMPALLPATHS: prints a summary of every min-sad-min path
found during a CONNECT or NEWCONNECT run to
file path.info. For each stationary point the energy, point group order and symbol,
Hessian eigenvalues and coordinates are given. Hessian eigenvalues are computed
for all stationary points. This min-sad-min format corresponds to the TRIPLES
keyword in the PATHSAMPLE program.
- DUMPDATA: creates a file in PATHSAMPLE min.data format for the minimum found
following a minimisation. Useful for a discrete path sampling initial path run to
create entries for the two endpoints.
- DUMPNEBEOS: creates the file EofS.neb for DNEB runs. This file is
also created if DEBUG is set; the default is false.
- DUMPNEBPTS: saves points for DNEB every iteration.
- DUMPNEBXYZ: creates the file neb.xyz for DNEB runs. This file is
also created if DEBUG is set; the default is false.
- DUMPPATH: prints a summary of the min-sad-min-
 -min path produced by CONNECT
or NEWCONNECT to
file path.info. For each stationary point the energy, point group order and symbol,
Hessian eigenvalues and coordinates are given. Hessian eigenvalues are computed
for all stationary points.
Note that the PATHSAMPLE program needs to be told if OPTIM is using
the DUMPPATH or DUMPALLPATHS format.
-min path produced by CONNECT
or NEWCONNECT to
file path.info. For each stationary point the energy, point group order and symbol,
Hessian eigenvalues and coordinates are given. Hessian eigenvalues are computed
for all stationary points.
Note that the PATHSAMPLE program needs to be told if OPTIM is using
the DUMPPATH or DUMPALLPATHS format.
- DUMPSP: specifies that information for all the stationary points located
should be dumped in
PATHSAMPLE min.data/ts.data format.
- DUMPVECTOR [ALLSTEPS] [ALLVECTORS]: if present the Hessian eigenvectors
will be written to file vector.dump after each OPTIMisation step,
preceded by the corresponding eigenvalue for each one. The
eigenvectors corresponding to zero eigenvalues are excluded. For hybrid eigenvector-following/BFGS
transition state searches only the results corresponding to the smallest non-zero
eigenvalue are dumped. The default is to dump only the vector corresponding
to the smallest non-zero eigenvalue for the last step. If ALLSTEPS is
present then vectors are dumped at every step. If ALLVECTORS is present
then all the vectors corresponding to non-zero eigenvalues are dumped, rather
than just one of them. ALLSTEPS and ALLVECTORS can be present in
either order.
- EDIFFTOL x: specifies the energy difference used to identify permutational isomers
in CONNECT and NEWCONNECT runs. Default is x
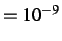 .
.
- EFIELD x: specifies that an electric field along the
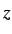 axis
should be included with magnitude xV/Å. For use with TIPnP potentials.
axis
should be included with magnitude xV/Å. For use with TIPnP potentials.
- EFSTEPS n: prints the unscaled steps in the Hessian
eigenvector basis every n steps--turned off by default.
- ENDHESS: calculate analytical Hessian and normal mode
frequencies at end of run. To obtain frequencies for UNRES you must
specify both ENDHESS and ENDNUMHESS.
ENDHESS is only intended for use in single geometry optimisations, and
should not be needed for CONNECT or PATH runs if DUMPPATH
is specified.
- ENDNUMHESS: calculate numerical Hessian and normal mode
frequencies at end of run.
Required if DUMPPATH or ENDHESS is specified for an UNRES run,
in which case it's an internal coordinate Hessian.
- ERROREXIT: indicates that the program should halt on error.
- EVCUT x: sets a cutoff value for eigenvalues. Eigenvalues
of magnitude less than
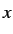 are treated as zeros and steps along such directions
are omitted.
are treated as zeros and steps along such directions
are omitted.
- EVOLVESTRING: use the evolving string method based on the
zero-temperature string method of E et al [15] for double-ended
transition state searches. Should be used in conjunction with the GROWSTRING keyword to set parameters.
- EXTRASTEPS x: obsolete; extra minimisation steps are allowed
for CHARMM runs proportional to the separation between two minima
multiplied by x.
- FAKEWATER: specifies that implicit water should be used in an AMBER
calculation via an effective dielectric constant.
- FILTH n: specifies an integer variable, n, to distinguish output files in
parallel landscape jobs. This keyword will be retired shortly in favour of the new
command line specification of file name extensions.
- FIXAFTER n: specifies that FIXIMAGE should be set permanently after step
n. This effectively freezes the interacting images in different supercells
for calculations with periodic boundary conditions.
- FIXATMS : For the GS and ES double-ended transition state search
methods, avoid overall translation and rotation by fixing all three
coordinates of one atom, confining another atom to a line, and a third atom
to a plane for all steps.
- FIXD t12fac dthresh: transition state search using hard sphere moves. A random
velocity vector is assigned and the first hard sphere collision is located. If t12fac
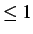 then the system is moved through a fraction t12fac of this first collision time; the
default value is 1. If
t12fac
then the system is moved through a fraction t12fac of this first collision time; the
default value is 1. If
t12fac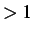 then the two atoms are moved half way between the entrance and exit of their
collision, but their separation is rescaled to unity. See also RANSEED.
Used in CONNECT procedure only if FIXD is set and the two
minima to be connected are closer than dthresh.
then the two atoms are moved half way between the entrance and exit of their
collision, but their separation is rescaled to unity. See also RANSEED.
Used in CONNECT procedure only if FIXD is set and the two
minima to be connected are closer than dthresh.
- FRACTIONAL: specifies fractional coordinates to be used in box length
optimisation for constant pressure runs using PV.
- FREEZE n1 n2 etc.: specifies that atoms n1, n2, etc. should be frozen.
Only works for LBFGS minimisation and BFGSTS if 3 or more non-linear atoms are frozen.
- FREEZENODES tol: For the double-ended transition state search
methods (NEB, GS, ES), at each step freeze those nodes for which the RMS
perpendicular force, as a fraction of the RMS force over all the images, is
less than tol. over all the images.
- GAMESS-UK system exec: potential will be generated by GAMESS-UK.
system is a string to identify the system and exec is
the name of an executable that will generate a GAMESS-US input deck from a points file.
If exec is omitted its name is assumed to be editit.system.
- GAMESS-US system exec: potential will be generated by GAMESS-US.
system is a string to identify the system and exec is
the name of an executable that will generate a GAMESS-US input deck from a points file.
If exec is omitted its name is assumed to be editit.system.
- GAUSSIAN: tells the program to read derivative information in
Gaussian92 format. This is unlikely to be working at the moment, since we
are not allowed access to the GAUSSIAN program.
- GDIIS x y z: initiates geometry direct inversion of the
invariant subspace. This option always seems to give poorer convergence
if analytic second derivatives are available at every step. It requires
three real parameters, which are read after the GDIIS keyword. The
first is the RMS force below which GDIIS will be used, the second is the
dimension of the DIIS problem to solve and the third is the interval between
GDIIS steps.
- GEOMDIFFTOL x: specifies the distance criterion used to identify identical isomers
in CONNECT and NEWCONNECT runs. Default is x
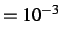 .
.
- GOT: Paul Whitford's Go model potential.
- GRADIENTS n: causes the gradients along the Hessian
eigenvectors to be printed every n cycles. Gradient printing is turned off by default.
- GRADSQ gsthresh nspecial nallow specifies optimisation of the modulus gradient
with nallow second order steps at intervals of nspecial
if the RMS force lies below gsthresh.
- GREATCIRCLE gcmxstp gcmxstp gcsteps gcconv: obsolete;
interpolation between two end points using a great circle in
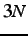 -dimensional
space.
-dimensional
space.
- GROWSTRING nimage iterd reparamtol growtol convtol mxstp:
specifies that double-ended transition state searches will be done using
either the evolving string method (based on that described by E et
al[15]) or the growing string method (based on that described by
Peters et al[16]). The default is growing strings; if the
separate keyword EVOLVESTRING is also specified, the evolving string
method will be used instead. nimage and iterd specify the
number of images and the maximal iterations per image to be used in the
first connect run; for all subsequent connections these parameters are
determined from the NEWCONNECT line. In the growing string case, the
iteration density affects only the maximum iterations allowed after the
string is fully populated with images. The string is reparametrised and the
images redistributed whenever the ratio of actual inter-image distance to
the desired distance falls below reparamtol for at least one
segment. For growing strings, a new image is added to one side of the
string whenever the RMS perpendicular force on one of the frontier images
falls below growtol. The string is deemed to have converged if the
RMS perpendicular force over all the images falls below convtol.
mxstp refers to the maximum allowed step for each image during the
optimisation. The default is a linear interpolation between image points;
use the CUBSPL keyword to switch this to a cubic spline interpolation.
- GSMAXTOTITD itd: Set the maximum total iteration density for the
growing string method. This specifies the maximum evolution iterations allowed per
total image number, including the iterations while the string is still
growing. If itd is less than 0, then this parameter is turned off
and there is no limit on the total iterations (this is the default).
- GUESSPATH gsteps maxgcycles gthreshold maxinte:
obsolete;
try to guess an interpolated path for sequential coordinate changes.
gsteps is the number of step to be tried for each sequential coordinate.
maxgcycles is the number of sweeps through pairwise exchanges
gthreshold is the coordinate change above which sequential changes are considered.
maxinte is the convergence criterion for the maximum allowed interpolated energy.
- GUESSTS guessthresh: use dihedral twisting in place of NEB for
transition state guesses with CONNECT for CHARMM and UNRES.
- HESSGRAD: For the growing string or evolving string double-ended
transition state search method, use the method described in the appendix of
Peters et al[16] to calculate the Newton-Raphson search
direction. Namely, the Hessian is approximated based on changes in the
gradient, and the tangential component of
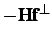 is projected
out. By default, the Hessian used is actually an approximation to the
derivative matrix of
is projected
out. By default, the Hessian used is actually an approximation to the
derivative matrix of 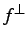 rather than the gradient.
rather than the gradient.
- HIGHESTIMAGE: obsolete;
only use the highest non-endpoint image in a double-ended MECCANO run.
- HUPDATE nstart ninterval phi: uses a Hessian update
procedure instead
of calculating the analytic second derivatives at every step.
nstart is the first step at which analytic derivatives will be calculated; by
default nstart=0, the Hessian is initialised to the identity and no
analytic second derivatives are ever calculated. ninterval is the interval
at which analytic Hessians are calculated; the default is 0, which means that no
additional analytic Hessians are ever found. A transition state search starting from
a minimum seems unlikely to work unless we calculate analytic Hessians at regular
intervals including the first step. Update applied is
phi times Powell[22] plus (1-phi) times Murtagh-Sargent.
(See Bofill and Comajuan, J. Comp. Chem., 11, 1326, 1995.)
- IH x: add an icosahedral field of strength x to the potential.
- INDEX n: initiates a search for a saddle of index n if
SEARCH 2 is specified. See also KEEPINDEX. Also works with BFGSTS
up to a maximum of index 50, but NOIT must be set and a Hessian is needed.
- INTEPSILON x: specifies the value of the
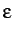 parameter
for diagonal shifts in the sparse internal coordinate transformation.
The default is
parameter
for diagonal shifts in the sparse internal coordinate transformation.
The default is 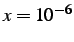 .
.
- INTMIN: specifies that internal coordinates be used in calls to mylbfgs,
including minimisation in the tangent subspace for hybrid eigenvector-following.
The algorithm used is that of Németh et al.[23]
Not recommended.
- KEEPINDEX : specifies that INDEX is set equal to
the number of negative Hessian eigenvalues at the initial point.
- LANCZOS : obsolete; specifies that a Lanczos procedure should be used to
find eigenvalues and eigenvectors.
Lapack routine DSYEVR is now used throughout instead.
- MACHINE: specifies that points should be saved in binary (machine) format instead
of human-readable xyz. Default false.
- MASS: specifies the use of mass weighting for the steps; the
output coordinates are not mass weighted.
- MAXBARRIER x: x specifies the maximum value for the smaller barrier height that
is allowed to constitute a connection during the
DIJKSTRA connection procedure.
- MAXBFGS x1 x2 x3 x4: x specifies the maximum allowed step length in LBFGS
minimisations, x1 for normal minimisations, x2 for Rayleigh-Ritz ratio
minimisation, x3 for putting structures in closest coincidence with
ministd, and x4 for NEB minimisations. Default values all 0.2.
- MAXERISE x1 x2: x1 is the maximum energy increase above which
mylbfgs will reject a proposed step. The default is x
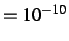 , but
a much larger value may be needed for CASTEP runs.
x2 performs the same function for the maximum rise in the Rayleigh-Ritz ratio
during minimisation in xmylbfgs.
, but
a much larger value may be needed for CASTEP runs.
x2 performs the same function for the maximum rise in the Rayleigh-Ritz ratio
during minimisation in xmylbfgs.
- MAXFAIL n: maximum number of failures
to lower the energy allowed in a minimisation before giving up.
- MAXGROWSTEPS n: For the growing string double-ended connection
method, specify a maximum number of steps allowed before another image is
added to the growing string. Default is 1000.
- MAXLENPERIM x: will stop the entire job if the total string
length for the growing strings or evolving strings method goes above x
times the total number of images. This usually means that something is going
wrong with the string. Default is 1000.
- MAXMAX x: specifies the maximum value that the maximum step size
is allowed to rise to. The default value is
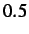 .
MAXMAX is also used as the uphill step size in hybrid eigenvector-following
transition state searches if the eigenvalue of the direction being followed is positive.
.
MAXMAX is also used as the uphill step size in hybrid eigenvector-following
transition state searches if the eigenvalue of the direction being followed is positive.
- MAXSTEP x1 x2: specifies the maximum step size for eigenvector-following and
steepest-descent calculations, default value 0.2. The precise
implementation of this constraint depends upon the step scaling method employed (see §8).
- MAXTSENERGY x: maximum ts energy that is allowed to constitute a connection during the
Dijkstra connection procedure. Paths are not calculated for transition states above
this threshold, and entries are not created in the path.info file.
Useful for CHARMM to ignore unphysical stationary points.
- MECCANO mecimdens mecmaximages mecitdens mecmaxit meclambda mecdist mecrmstol
mecstep mecdguess mecupdate: obsolete; an interpolation between
end points via rigid rods of variable length.
- MINMAX x: specifies the minimum value that the maximum step size
is allowed to fall to. The default value is
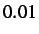 .
.
- MODE n1 n2: specifies the eigenvector to be followed uphill in an eigenvector-following
transition state search,
where 1 means the softest mode, 2 means the next softest, and so on.
Setting n1 to 0 means that the softest mode is followed uphill at every step,
otherwise a maximum overlap criterion is used to determine the mode followed after the
first step. The sign of n1 is used to determine the direction in which we
step along the eigenvector if we are starting from a stationary point. This provides a way
to start transition state searches from minima along any of the eigendirections in
either sense. If a minimisation is started from a converged transition state then setting
n1 to
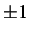 enables the program to walk down the two sides of the pathway. This is true for
runs based on LBFGS techniques too if the keyword BFGSSTEP is used.
MODE is now also used in EF/LBFGS searches so long as an analytical Hessian is
calculated. n1 is used for the first step and n2 for subsequent steps. The
default is 0 for both parameters. Note that n2 must be explicitly set equal to
n1 to follow the same mode (as judged by overlap) in following steps.
enables the program to walk down the two sides of the pathway. This is true for
runs based on LBFGS techniques too if the keyword BFGSSTEP is used.
MODE is now also used in EF/LBFGS searches so long as an analytical Hessian is
calculated. n1 is used for the first step and n2 for subsequent steps. The
default is 0 for both parameters. Note that n2 must be explicitly set equal to
n1 to follow the same mode (as judged by overlap) in following steps.
- MORPH morphmxstp mnbfgsmax1 mnbfgsmax2 morphemax morpherise msteps:
attempt to morph between endpoints by taking steps either towards or away from the
endpoint specified in finish, and minimising in the tangent space.
morphmxstp is the maximum step size.
mnbfgsmax1 is the maximum number of minimisation steps taken in the tangent
space when the distance from the finish endpoint is greater than the
step size convergence criterion.
mnbfgsmax2 is the maximum number of minimisation steps taken in the tangent
space when the distance from the finish endpoint is less than the
step size convergence criterion.
morphemax is the maximum energy allowed during the interpolation;
if this threshold is exceeded the algorithm toggles between stepping away/towards
the finish endpoint.
morpherise is the maximum energy rise permitted in a step.
msteps is the maximum number of MORPH steps allowed.
Used successfully with the general BLN model for protein L, where linear
interpolation leads to strands crossing.
- MOVIE: a movie will be dumped in xyz format for the AMBER potential.
- MSEVBPARAMS shellsToCount maxHbondLength minHbondAngle OOclash_sq:
specifies parameters for the MSEVB potential.
- NATB: specifies a tight-binding potential for sodium, silver and lithium.
- NEB nsteps nimage rmsneb: specifies a nudged elastic band pathway
calculation[24,25] using a maximum of nsteps steps with
nimage images and RMS convergence criterion rmsneb. The default values are
nsteps=100, nimage=10, rmsneb=0.1.
- NEWCASTEP system: tells the program to use the new CASTEP
executable to calculate energies and
forces with input files based on the system name system. Periodic boundary
conditions are assumed, so projection is not performed to remove overall rotation.
- NEWCASTEPC system: tells the program to use the new CASTEP to calculate energies and
forces with input files based on the system name system.
In this case the system is assumed to be a cluster, so that both translational and rotational
degrees of freedom correspond to zero Hessian eigenvalues at a stationary point.
- NEWCONNECT NConMax NTriesMax ImageDensity IterDensity ImageMax ImageIncr:
specifies the new connection algorithm described by Trygubenko and Wales.[14]
A finish file containing the coordinates of a second minimum is required in the
current working directory, as for CONNECT.
NConMax is the maximum number of cycles allowed, default=100.
NTriesMax is the maximum number of connection attempts for any pair of
local minima in the stationary point database, default=3.
ImageDensity is a real variable specifying the number of DNEB images to be used for
the first connection attempt for any pair of minima, default=10.0.
An image density is specified, so that the actual number of images is ImageDensity
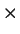 the
separation between the two minima.
If NTriesMax
the
separation between the two minima.
If NTriesMax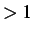 then
the image density is increased by ImageIncr for each attempt, up to
a maximum of ImageMax images in total.
The default values for ImageIncr and ImageMax are 0.5 and 400, respectively.
IterDensity specifies the maximum number of iterations used to optimise the
DNEB in terms of a density. The actual value used is IterDensity
then
the image density is increased by ImageIncr for each attempt, up to
a maximum of ImageMax images in total.
The default values for ImageIncr and ImageMax are 0.5 and 400, respectively.
IterDensity specifies the maximum number of iterations used to optimise the
DNEB in terms of a density. The actual value used is IterDensity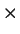 the
separation between the two minima. The default value of IterDensity is 30.0.
the
separation between the two minima. The default value of IterDensity is 30.0.
- NEWNEB Nimage NIterMax RMSTol:
specifies that the DNEB algorithm[14]
should be used for a double-ended transition state search.
Nimage, NIterMax and RMSTol are the number of images, the maximum
number of iterations and the RMS force convergence condition, respectively.
The default value for Nimage is 2 and there are no defaults for the other parameters.
If NEWCONNECT is also specified then these values are used for the first NEWCONNECT
cycle only. However, if CONNECT is specified they are used for every cycle.
- NGUESS n: n is the number of transition state guesses to be tried in
guessts for CHARMM or UNRES before switching back to NEB or NEWNEB.
- NOCISTRANS minomega: used in CONNECT runs to reject transition states
that connect two minima with different omega angles, i.e. to prevent cis-trans peptide
isomerisation. Checks that all residues besides proline are in the
trans state and rejects transition states and minima that are not,
i.e. with
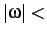 minomega. minomega defaults to 160 degrees.
For proline every
minomega. minomega defaults to 160 degrees.
For proline every 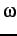 is allowed.
is allowed.
- NOHESS: no Hessian should be calculated during geometry optimisation.
Hybrid EF/LBFGS transition state searches are therefore conducted without a
Hessian. The smallest eigenvalue and the corresponding eigenvector are found from a separate
Rayleigh-Ritz minimisation problem for every combined EF/BFGS step.
- NOIT: for use with hybrid eigenvector-following runs when a Hessian is available.
Specifies that the required lowest eigenvalue and eigenvector be found using
LAPACK routine DSYEVR instead of by iteration.
- NOLBFGS: For the growing string or evolving string double-ended
transition state search methods, instead of using L-BFGS optimization to
evolve the strings, simply take steps in the direction of the perpendicular force.
- NONEBMIND: do not put end point structures supplied to DNEB into
closest coincidence.
- NONLOCAL x y z: factors for averaged Gaussian, Morse type 1 and Morse
type 2 potentials.
- NOPOINTS: printing of coordinates to file points for intermediate steps
is turned off. The default is true.
- NORANDOM randomcutoff: used in CHARMM transition state guessing procedure
together with TWISTTYPE. Setting randomcutoff very large prevents random
steps, and is recommended.
- NORESET: specifies that periodic images should not be returned to the
primary supercell for calculations involving periodic boundary conditions. (May be
useful for calculating transport properties with kinetic Monte Carlo).
- NOTE: has the same effect as COMMENT, i.e. the rest of the line
is ignored.
- ODIHE: for use with CHARMM. Specifies calculation of
a dihedral-angle order parameter using the reference structure supplied in ref.crd.
- OH: add an octahedral field of strength x to the potential.
- OPTIMIZETS: optimise transition states at the end of each DNEB run.
Default false. Set to true by NEWCONNECT.
- ORT: remove overall rotation and translation for DNEB runs. Default false.
- OSASA rpro: for use with CHARMM. Specifies calculation of
a solvent accessible surface area order parameter using probe radius rpro.
- PARALLEL nproc: specifies that auxiliary programs such as GAMESS, CASTEP and
CPMD use nproc processors.
- PARAMS: enables additional parameters to be set for
specific potentials, e.g.
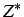 for LJAT,
for LJAT, 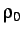 for Morse, n, m, the
box lengths
for Morse, n, m, the
box lengths 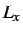 ,
, 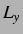 and
and 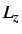 and the cutoff for ME, the
box lengths
and the cutoff for ME, the
box lengths 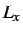 ,
, 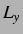 and
and 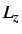 and the cutoff for JM, SC and P6,
and
and the cutoff for JM, SC and P6,
and 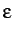 ,
, 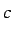 and
and 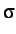 for Au, Ag and Ni. Note that all the
numbers are read as real.
for Au, Ag and Ni. Note that all the
numbers are read as real.
- PATH n: requests calculation of the pathway connecting two minima from the transition
state specified in odata. n is the number of intermediate points files to save on either
side, default n=3.
To choose which frames are dumped in the path.xyz file a stochastic method is employed.
All three stationary points are always included.
Other frames are dumped with a probability proportional to n.
If n exceeds the number of frames in the path then they will all be dumped, up to a
maximum of 100.
The complete xyz file is printed to path.xyz and the energy as a function of
path length is printed to file EofS. A summary of the path characteristics such as barrier heights,
path lengths and cooperativity indices is printed at the end. If READVEC is present in
odata then the required eigenvector corresponding to the displacement direction will
be read from vector.dump, otherwise it will be calculated by diagonalizing a Hessian (for
SEARCH 2) or using an iterative approach (for BFGSTS with or without
NOHESS; NOIT can also be used with BFGSTS.)
After the initial step the energy will be minimised by whichever first or second
order method is specified in odata, i.e. eigenvector-following or second-order Page-McIver
for SEARCH 0 and 6 and 7, LBFGS for BFGSMIN, and first order steepest-descent
for BSMIN and RKMIN. See also DUMPPATH.
- PERMDIST: minimise distances between stationary points with respect to
permutational isomerisation.
Requires the auxiliary file perm.allow to specify permutable atoms, otherwise
all atoms are assumed to be permutable. The absence of a perm.allow
file is considered a mistake for CHARMM runs.
The first line of the perm.allow file must contain an integer
that specifies the number of groups of interchangeable atoms.
The groups then follow, each one introduced by a line with two integers
that specify the number of permutable atoms and the number of other pairs
of atoms that must swap if the first pair are permuted.
The following line contains the numbers of the permutable atoms and then
the numbers of the swap pairs, in order.
For CHARMM runs the perm.allow file can be generated automatically
from the input.crd file using the python script perm.py
written by Dr Mey Khalili.
Note that it is then essential to use CHARMM topology files
that include symmetrised definitions of all the relevant amino acids.
- PERTDIHE chpmax chnmin chnmax iseed: CHARMM
and UNRES dihedral angle perturbation specification.
Performs random, GMIN-style twists before starting optimisation.
- POINTS: this is usually the last keyword and introduces the
coordinate information, which is read one atom per line. Each line must consist
of the atomic symbol of the atom followed by its three Cartesian coordinates.
Leading spaces before the atomic symbol should be avoided. For calculations
involving CHARMM, UNRES, AMBER and CPMD the coordinates are obtained from auxiliary files,
and POINTS will be the last OPTIM directive in the odata file, or
is not used (CHARMM and UNRES).
- PREROTATE: For the GS and ES double-ended transition state
search methods, if using FIXATMS to zero some coordinates of the
forces to avoid overall translation and rotation, this keyword will rotate
the start and end points so that those coordinates are zero in both.
The default is true.
- PRESSURE: if present
tells the program to perform a constant zero pressure optimisation
for bulk SC, ME, MP, MS and P6. Normally a constant volume optimisation takes place.
- PRINT n: sets the print level. The default value is zero. Use
of this option is not recommended, since more specific print control is possible.
- PRINTCOEFFICIENTS: prints ground state coefficients for the MSEVB potential.
- PUSHCUT x: sets the threshold for the RMS force below which the
system may apply a PUSHOFF to escape from a stationary point of the wrong Hessian
index. Default value is 0.00001. Using a negative value prevents pushoffs from
occurring.
- PUSHOFF x: sets the magnitude of a step away from a stationary
point of the wrong Hessian index (see §8 for default action). Default value is 0.01.
Formally used as the step size in hybrid eigenvector-following
transition state searches if the eigenvalue of the direction being followed is positive.
MAXMAX has now replaced PUSHOFF in this role.
- PV press gtol grad steps: specifies an approximate constant pressure optimisation with
pressure press and convergence criterion gtol for the gradients with
respect to the box lengths. If any box length gradient is larger in magnitude than
grad then up to a maximum of steps optimisation steps for the box lengths
are performed.
If the three box lengths are equal to start with and the keyword CUBIC is included,
then a cubic box is maintained.
Will not work with atom types MP, ME and MS.
Default values, press=0, gtol=0.001, grad
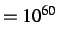 , steps=100.
, steps=100.
- PVTS press gtol grad nboxts: as for PV but searches for a transition
state with respect to box length nboxts, where nboxts=1, 2 and 3 specify the
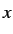 ,
, 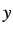 and
and 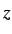 box lengths, respectively. nboxts defaults to 1.
box lengths, respectively. nboxts defaults to 1.
- RADIUS x: specifies that atoms should not be allowed to move beyond a
distance x from the origin.
- RANSEED n: specifies the initial seed for random number generation, as used in
hard sphere moves with FIXD.
- RATIOS: specifies that the pathway parameters needed to calculate catastrophe
ratios should be printed in a PATH run.
- READPATH: if specified with CALCRATES then rate constants will be
calculated for the transition states found in path.info and no stationary point
searches will be performed.
- READSP: instructs OPTIM to read minima and
transition state data in the PATHSAMPLE format.
This has not been implemented yet!
- READHESS: Hessian will be read from file derivs at first step.
- READVEC: if present the Hessian eigenvector
corresponding to the smallest eigenvalue
will be read from file vector.dump. The latter file can be generated using the
DUMPVECTOR keyword in a previous run.
- REDOPATH: instructs OPTIM to read transition state coordinates
successively from the file redopoints during a NEWCONNECT run.
Can be used to generate movie files from stationary point sequences calculated
by PATHSAMPLE. The correct start and finish coordinates for the minima
must be present in the odata and finish files.
- REDOPATHXYZ: as for REDOPATH above, except that existing
path.n.xyz files are read instead of actually calculating pathways for the
transition states.
- REOPT: specifies that the eigenvector to be followed should be reoptimised
in a BFGSTS search after the EF step and before the tangent space minimisation.
This is probably not a good idea. Default is false.
- REOPTIMISEENDPOINTS: specifies that endpoint minima should be reoptimised
if their RMS force lies above the convergence threshold. Useful in connection
runs where failure is guaranteed if one or more of the endpoints is not identified
as a minimum. A large RMS force at the beginning of a connection run driven
by PATHSAMPLE should be investigated because it suggests a problem, e.g. with
CHARMM permutational isomerisations.
Default is false.
- REPELTS: specifies that the transition state search direction should
be orthogonalised to the coordinates in file points.repel.
- RESIZE x: scales all the coordinates by x
before the first step is taken.
- RKMIN gmax eps: calculates a steepest-descent path using gradient only
information with convergence criterion gmax for the RMS force and initial
precision eps. A fifth order Runga-Kutta algorithm is used.
- ROT: if present a rotational kinetic energy term is
added to the Hamiltonian. Either constant angular momentum (ROT JZ x) or
constant angular velocity (ROT OMEGA x) may be used to specify
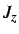 or
or 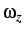 .
.
- SCALE n: sets the scaling algorithm for eigenvector-following
steps, as described in §8. The default is n=10.
- SCORE_QUEUE: specifies an Score parallel environment.
Currently not used on group clusters.
- SEARCH n: specifies the search type for eigenvector-following and
steepest-descent calculations based on second derivatives and Hessian diagonalisation, default is type 0.
The most common options are 0, a minimisation, and 2, a transition state search. See
§5 for full details.
- SHIFT x: specifies the shift applied to eigenvectors corresponding
to normal modes that conserve the energy. The default is
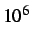 . The shift must be
large enough to move the eigenvalues in question to the top of the spectrum.
. The shift must be
large enough to move the eigenvalues in question to the top of the spectrum.
- SQVV NIterSQVVGuessMax SQVVGuessRMSTol:
specifies that the first NIterSQVVGuessMax DNEB refinements should be
done using the SQVV algorithm, rather than LBFGS.[14]
The DNEB optimisation will switch to LBFGS minimisation after NIterSQVVGuessMax
steps or if the RMS force falls below SQVVGuessRMSTol.
The default values for NIterSQVVGuessMax and SQVVGuessRMSTol
are 300 and 2.0, respectively.
- STEPMIN n: sets the minimum number of optimisation steps before convergence
will be tested. The default is zero.
- STEPS n: sets the maximum number of optimisation steps per call
to OPTIM. The default is n=1. Setting n to zero at the start of a run
means that only a point group analysis is performed.
- STOPB: specifies an alternative stopping condition, designed for use with
the PATHSAMPLE program. OPTIM will read in data from a min.B file if it exists.
CONNECT runs will stop if a minimum in min.B is found.
- STOPDIST x: in a CONNECT run, stop as soon as the distance between
the initial (x
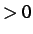 ) or final (x
) or final (x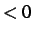 ) minimum and the furthest connected minimum
exceeds
) minimum and the furthest connected minimum
exceeds 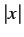 .
.
- STOPFIRST: in a CONNECT run, stop as soon as the initial minimum has a transition state
connection.
- SUMMARY n: prints a summary of the steps and derivatives
every n steps. The default is
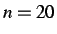 .
.
- SYMCUT x: specifies the RMS force below which the point
group symmetry subroutine will be called. The default is x=0.001.
- TAG num massfac:
allows atoms to be tagged. Atom number num has its mass increased
by a factor massfac for point group symmetry assignment only.
To tag more than one atom use additional TAG lines in odata.
- TD: add a tetrahedral field of strength x to the potential.
- TOLD x: sets the initial distance tolerance for points
to be considered the same after rotations and reflections in point
group determination. The default is 0.0001.
- TIMELIMIT x: OPTIM will stop if the accumulated cpu
time exceeds x seconds. The default value is infinity.
- TOLD x: initial distance tolerance in symmetry subroutine.
The default is 0.0001.
- TOLE x: sets the initial tolerance for eigenvalues
of the inertia tensor to be considered equivalent in point
group determination. The default is 0.0001.
- TOMEGA: includes omega angles in the TWISTDIHE list.
- TOSI
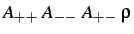 : specifies a Born-Mayer potential
with the parameters indicated (see §4).
: specifies a Born-Mayer potential
with the parameters indicated (see §4).
- TOSIC6 c6pp c6mm c6pm: specifies additional C6 dispersion coefficients
for the TOSI potential. c6pp, c6mm, and c6pm should be the values of the C6
parameters between positive ions, negative ions and positive and negative ions in atomic units
(see §4).
- TOSIPOL alphap alpham damp: specifies cation and anion polarisabilities in atomic units
and a damping parameter for the TOSI potential (see §4).
- TRAD x: sets the trust radius (see §8). The default value is 2.0.
- TSIDECHAIN: includes sidechain angles in the TWISTDIHE list.
- TWISTDIHE nmode xpert: twist phi/psi dihedral angle nmode by
xpert degrees before starting optimisation.
- TWISTTYPE n: integer n specifies the type of dihedral angle twisting
used to guess transition states in guessts for CHARMM and UNRES.
The recommended value is n=5. See also NORANDOM.
- TWOENDS fstart finc ntwo rmstwo ntwoiter twoeval: obsolete: specifies a double-ended transition
state search. fstart is the initial force constant pulling the coordinates towards the final
geometry which is read from a file named final. finc is the increment for the force
constant after each cycle. ntwo is the maximum number of minimisation steps taken after each
increment of the force constant, and rmstwo is the convergence condition on the RMS force
for these minimisations. ntwoiter is the total number of force constant increments allowed
and twoeval is the value of the smallest Hessian eigenvalue below which the program will
shift to a transition state search.
- UNRES: specifies the UNRES potential, which requires an UNOPTIM executable.
An auxiliary file coords is required. UNRES must be the last OPTIM directive in the
odata file. The remaining content of odata consists of UNRES keywords and
setup information.
- UPDATES mupdate1 mupdate2 mupdate3 mupdate4:
specifies the number of updates in the LBFGS routine before
resetting.
mupdate1 is used for energy minimisation,
mupdate2 is currently ignored by mind,
mupdate3 is used in Rayleigh-Ritz eigenvalue calculations, and
mupdate4 is used in NEB and DNEB optimisation.
mupdate5 is used in string method optimisation.
The default value is 4 for all five parameters.
- VALUES n: prints the Hessian eigenvalues every n steps.
The default is n=20.
- VARIABLES: optimises a general function (which must be specified in
potential. The initial values of the variables are read one per line after the POINTS
keyword in odata.
- VECTORS n: prints the Hessian eigenvectors every n
steps. Eigenvector printing is turned off by default.
- WARRANTY: print warranty information.
- WELCH
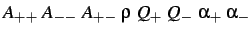 : specifies a Welch binary
salt potential with the parameters indicated (see §4).
: specifies a Welch binary
salt potential with the parameters indicated (see §4).
- ZEROS n: sets the number of zero Hessian eigenvalues to be
assumed--useful for general optimisation problems specified by VARIABLES.


Next: Specification of Potentials
Up: OPTIM3 User Guide
Previous: The odata file
David Wales
2007-06-07