- ADDMIN file: reads in the data for one or more minima from file
in OPTIM min.data.info format and then stops.
Any number of min.data.info files can be concatenated into file.
New entries will be created in min.data and points.min.
If min.A and min.B do not exist they are not created.
READMIN therefore provides an alternative way to start an initial
path calculation by providing minima, so long as min.A and min.B
are created by hand.
It also provides a way to add additional minima to an existing database, since
new entries should be appended. There is a check to make sure that the minima
added are different from all those currently known.
This keyword has the same function as READMIN and is provided for convenience.
- ADDMINXYZ file: reads coordinates in xyz format from
file file and runs OPTIM for each one using odata.addminxyz,
which needs to contain the OPTIM keywords DUMPDATA and ENDHESS
so that a min.data.info file is created.
The resulting min.data.info files are then used to add minima to the
database, which can be empty initially. CYCLES must also be set to a non-zero value.
- ADDPATH afile : reads pathway data from file afile.
Up to 1000 ADDPATH lines are allowed in pathdata.
If ADDPATH is used on its own before a connection has been
achieved then DIJINITCONT should be specified so that the
pairdist and pairlist files are written is required. See also INITIALDISTANCE.
- ADDPERM: adds permutational isomers of every
stationary point to the databases. Not recommended except for small systems
where you are using a tagged atom to study an isomerisation (e.g. 2D LJ7).
- ADDPERM2 start finish: adds all distinct permutation-inversion isomers of every
stationary point to the databases for permutations of atoms numbered from
start to finish, inclusive. Not recommended except for small systems!
Added this capability to allow studies where permutation-inversion isomers are
not automatically lumped together, to see the effect of free energy regrouping
(or observation time scale) on the landscape.
Any perm.allow file present is ignored.
- ADDPERM3: adds all distinct permutation-inversion isomers of every
stationary point to the databases according to the allowed permutations in
perm.allow.
Can be used in combination with MERGEDB to create a new database where
permutation-isomers are distinguished.
- ADDPERMSCF : adds sign permuted copies of the solutions found in min.data and ts.data, respectively. This keyword is specifically used with the hybrid version of OPTIM with QChem.
- ALLOWAB: allows minima to belong to both A and B sets where lumping
is performed with REGROUPFREE and RFMULTI.
This may be useful for understanding how free energy groups merge as a function of
observation time scale and temperature, when rate constants are not required.
- ALLTS : specifies that transition states that are considered identical by energy
and geometry conditions to one already in the database, but which connect different minima, are not discarded
and are treated as a new transition state. Different connections for the same transition state can occur because
of numerical noise in the pathway calculation, and it is possible to overlook the correct connection unless
ALLTS is set.
- ANGLEAXIS : specifies angle-axis coordinates for rigid bodies. NB: this is not part of the GENRIGID framework.
- AMBER9 : specifies the AMBER 9 potential.
- AMBER12 : specifies the AMBER 12 potential.
- AMH : specifies an AMH potential.
- AMHQ whichmin : Calculates Wolynes Q score based on the native state structure.
- AMHRMSD whichmin : Calculates RMSD based on the native state structure.
- AMHQCONT whichmin rcut : Calculates Q-cutoff score based on a native distance.
- AMHRELQ whichmin1 whichmin2 : Calculates Wolynes Q score between two minima.
- AMHALLATOM : Adds sidechains and completes backbone for AMH structures with SCWRL.
Input structure is amhmin.pdb. Output structure is amhmin.pdb.scwrl. Scwrl output logs scwrl_out.
- AMH_RELCO whichmin rcut : Calculates relative contact order for AMH structures.
- ARNOLD: call the Lanczos routines from ARPACK to perform eigendecomposition associated
with the WATPDFSYM keyword.
- ARNOLDI: call a weighted Arnoldi scheme to diagonalise the full rate matrix.
- ATOMMATCHDIST: Currently for bulk systems only, providing an alternative method for finding the shortest distance between structures.
This method is particularly useful for finding the shortest distance between very similar structures such as defective crystal structures.
The method is an extension of the original bulkmindist.f90 routine for matching permutational isomers. Atoms are overlayed and the number of exactly matching atoms is maximised.
With ATOMMATCHDIST, the method is non-deterministic to maximise efficiency. It has been optimised to work particularly well for providing small distances
between similar structures and for removing very large distances. However, it will not always outperform the default method for middling distances.
Currently only one method for calculating distances, atom matching or the default, can be used.
- ATOMMATCHFULL: As for ATOMMATCHDIST, but provides a slow deterministic result that should always outperform or equal the default distance calculation and ATOMMATCHDIST but may never finish.
- BHINTERP dthresh maxe bhsteps conv T stepsize accrat K sfrac ICINTERP: specifies that
OPTIM jobs should be run using only the BHINTERP interpolation option,
where no transition state searches are run. Requires the auxiliary file odata.bhinterp.
This option is intended for use with DIJINITSTART and DIJINITCONT.
The various arguments are used to add a BHINTERP line to the OPTIM odata file.
The distance threshold parameter specifies that interpolation between consecutive minima
in the Dijkstra list should occur if their minimum distance is greater than dthresh.
The threshold actually used in the corresponding odata file is either the minimum distance
divided by two or dthresh, whichever is greater. This arrangement enables
parallel OPTIM jobs to be run for different pairs of minima.
Intermediate minima are only accepted if their energy is below maxe.
A basin-hopping global optimisation run of bhsteps is run for each pair of
end point minima within dthresh using an RMS gradient convergence criterion
of conv, a temperature parameter of T, and a maximum step size for
perturbations of stepsize. The step size is adjusted dynamically towards an
acceptance ratio target of accrat.
The objective function consists of the energy of the minimum on the potential energy surface
plus the energy corresponding to harmonic springs of force constant K
stretched to displacements corresponding to the minimum distance between the new minimum
and the end points.
sfrac is used in the initial interpolation: a value of 0.5 will put the initial
guess half-way between the end points, and in general the geometry will be sfrac times one
end point plus (1 -sfrac) times the other.
For large distances, using a value other than a half may be helpful.
If ICINTERP is present then amino acid side chains are interpolated using
internal coordinates for CHARMM runs.
The step size is interpreted in degrees for CHARMM, where the perturbations used
to step between minima are performed using dihedral angle twists.
The algorithm is applied recursively between minima as new minima are found.
A new minimum will not be accepted if both distances to the minima we are
currently trying to interpolate between are greater than the minimum distance
between these minima.
- BISECT dthresh maxe bisectsteps attempts ICINTERP: specifies that
OPTIM jobs should be run using only the BISECT interpolation option,
where no transition state searches are run. Requires the auxiliary file odata.bisect.
This option is intended for use with DUMMYTS.
The various arguments are used to add a BISECT line to the OPTIM odata file.
The distance threshold parameter specifies that bisection between consecutive minima
is attempted if their minimum distance is greater than dthresh.
The interpolated geometry is minimised and compared with the starting structures.
New minima are only accepted if their energy is below maxe; they are added to the
current list of known minima in the OPTIM job between the two starting structures.
The estimated energy of any dummy transition states between the two starting structures
is raised if the sum of distances between the new minimum and the two structures is
greater than the original minimised distance.
bisectsteps is the maximum number of bisection steps, and
attempts is the number of attempts per step for a given pair.
If minimisation leads to one of the end points the interpolation is changed to use
fractions of the two structures that become increasingly skewed, as for
the adjustment of sfrac with BHINTERP, above.
The ts.data file is rewritten if the any dummy transition state energies are revised.
If ICINTERP is present then amino acid side chains are interpolated using
internal coordinates for CHARMM runs.
BOXDERIV: specifies a periodic system with a variable cell. The cell parameters (nine components
for three cell vectors) will be saved in binary files cell.min and cell.ts.
They are communicated to PATHSAMPLE from OPTIM in the pathinfo and
min.data.info files, and from PATHSAMPLE to OPTIM using three lines
at the end of start and finish files.
The coordinates are all saved as fractional; minimum vectors can be obtained
for alignment using BULK 1.0 1.0 1.0, which sets the three box lengths to one
in the orthorhombic minimum image formulation.
See also BOXISOMERTOL.
- BOXISOMERTOL svd vol: checks for identical isomers in systems with variable cells when
BOXDERIV is in use. svd and vol are the tolerances for a singlevalue decomposition based on
a distance matrix and for the cell volume. For stationary points to be considered permutation-inversion isomers
the energy difference must also be lower than the EDIFFTOL value.
See also BOXDERIV.
- BULK x1 x2 x3: a bulk system in a periodic box of lengths x1, x2, x3.
- CALCORDER: the program will call subroutine calcorder.f90 and calculate
order parameters, before exiting. Since the order parameters will be system dependent
users must compile in an appropriate calcorder.f90 themselves.
- CAPSID rho eps rad h: parameters for a rigid pentagonal pyramid. If h is omitted
it is set to half rad by default.
- CHARMM: specifies that the CHARMM potential is used in the OPTIM runs.
- CHECKCONNECTIONS: instruction to check that each minimum has at least the number
of connections specified by the CONNECTIONS keyword at the start of every run,
before anything else is done. See CONNECTIONS below.
This keyword results in calls to the original single-ended transition state
searching subroutine, which is not efficiently parallelised.
The NEWCONNECTIONS keyword should probably be used instead.
- CHECKMETRIC: check the distance metric for the Dijkstra missing connections path
in runs with INITIALDISTANCE. There should be a transition state connecting pairs with
zero distance, and the distance should be less than or approximately equal to the value previously
calculated if there is no direct connection yet.
- CHECKMIN start finish: will rerun OPTIM for all the
minima from start to finish using the odata file
odata.checksp, which must be present in the working directory.
PATHSAMPLE will report whether or not the OPTIM job converged, so the number
of minimization steps should be set accordingly (e.g. BFGSSTEPS 1).
This option may be useful for finding any unconverged minima in a corrupted database,
which could then be removed using REMOVESP. See also DIJCHECKMIN.
- CHECKTS start finish: will rerun OPTIM for all the
transition states from start to finish using the odata file
odata.checksp, which must be present in the working directory.
PATHSAMPLE will report whether or not the OPTIM job converged.
This keyword is useful for recreating path.info files for transition states, perhaps
after changing the potential, and hence relaxing the original transition state.
Such runs will usually use BFGSTS in OPTIM, with a PATH keyword to
run the corresponding pathway, and DUMPALLPATHS to produce the path.info file.
odata.checksp could be the same as for CHECKMIN, as in, a minimization
with zero steps where only the initial energy and gradient are calculated to test for convergence
and the Hessian index is NOT checked.
This option may be useful for finding any unconverged transition states in a corrupted database,
which could then be removed using REMOVESP.
- CHECKTSFILE tsfile: will rerun OPTIM for all the
transition states listed in the file tsfile using the odata file
odata.checksp, which must be present in the working directory.
See CHECKTS above and also DIJCHECKTS.
- CHECKPATHINFO: will attempt to read any path.info.n files in the current
working directory, where n is an integer. These files will be renamed with a .done
ending after processing. Hence it is possible to automatically read path.info.n files
from a previous PATHSAMPLE job that died for some reason.
This check is now run on setup and after every OPTIM job finishes. Hence it is now
possible to communicate path.info files between PATHSAMPLE jobs running in different places.
- CHECKSP_MUT: invokes the CHECKSPMUTATE subroutine. To be used in conjunction
with CHECKMIN or CHECKTS. Mutations can therefore be made to the stationary points before
reoptimisation. Auxiliary files nresidueslog, resnumberlog, atomnumberlog,
oldreslog, newreslog and original_protein.pdb are required. Also requires
NATOMS_NEW KEYWORD if the number of atoms in the system changes with the mutation, and
NATOMS_CHAIN if the system has non-protein atoms.
- CLOSEFILES: open and close the min.data and ts.data files as needed.
By default these files stay open all the time. For small systems this inevitably seems to cause
file corruption on nfs mounted file systems, which is apparently due to bugs in the interaction
of nfs with the Linux kernel. The symptom is that strings of control characters appear randomly in
min.data and/or ts.data, which means that the run cannot continue without
manually fixing the file in question.
The CLOSEFILES keyword has provided a successful workaround for this problem.
CLOSEFILES is now set to TRUE by default. There is no need to set the keyword. 7/6/2024.
- CONNECTIONS n maxattempts: minimum number of connections for each minimum.
For each new minimum found PATHSAMPLE will attempt to find n connected minima.
If n is one or fewer then no additional single-ended transition state searches are
needed, and the more efficient cycle2.f90 subroutine is used. In this case, OPTIM
jobs do not have to be processed in batches; instead, a new OPTIM job is submitted
as soon as one finishes. The default for n is now 0.
The maximum number of transition state searches from each new minimum is maxattempts,
for which the default is 10.
Note that you will need file odata.tssearch in the current
working directory if n is greater than one.
The odata.tssearch file needs to contain OPTIM PATH
and DUMPALLPATHS directives
so that a path.info file is created.
It is no longer necessary to supply a separate odata.path file.
This keyword results in calls to the original single-ended transition state
searching subroutine, which is not efficiently parallelised.
The NEWCONNECTIONS keyword should probably be used instead.
- CONNECTPAIRS connectfile: minima are chosen for connection attempts based
on the pairs specified in file connectfile, which must have two integers per line
corresponding to the order in min.data.
- CONINT specifies that terms for internal minimum distances
should be included in the energy and gradient for the auxiliary interpolation
potential when INTCONSTRAINT is specified.
- CONNECTREGION min1 min2 dist: grows the stationary point database by running
double-ended transition state searches between minima whose minimum separation is less then
dist. min1 and min2 specify the starting minima in the double loop over
pairs, so that restarted runs need not consider pairs that have previously been searched.
Minima that already have a direct connection are ignored.
The maximum number of candidate pairs calculated per call to connectd.f90 is 10000.
This parameter may be useful for exhaustive searches for small systems.
- CONNECTUNC option (refmin): attempt to connect minima not connected to the AB region (unconnected minima).
option specifies one of three options (no default set): LOWEST (n) attempts connections of the lowest
energy unconnected minima with the n closest minima that belong to the set
connected to A and B, EREF refmin attempts connections
of the minimum refmin with the minima closest in energy, DISTREF refmin does the same using
the closest minima in distance to refmin.
Note that EREF and DISTREF will connect to other minima that may themselves be unconnected.
To attempt connections to the closest minima in the connected set use
DISTREFAB.
There is one other option LOWESTTEST that runs the LOWEST
version and then exits after printing the selected minima.
- COPYFILES a1 a2 a3 …: specify additional files that need to be
copied to distributed nodes. Only files odata.<pid> and finish.<pid> are
copied by default. For example, the BLN potential requires file BLN sequence, while
CHARMM runs may need one or more crd files. Each string must be less than or equal to
twenty characters in length, and the total, including separating blanks, must not exceed
250 characters. If the CHARMM keyword is set and the COPYFILES
arguments do not include input.crd then the program will stop.
- COPYOPTIM: if present, some of the OPTIM output files will be copied to
the mother superior node and not deleted. This behaviour also occurs if the DEBUG
keyword is present. The copied files, in addition to path.info (min.data.info
for a BHINTERP run), are the OPTIM output file, the odata file
and the finish file.
- CPUS ncpu: specifies the number of cpu's (cores) available when running
PATHSAMPLE interactively, without using a scheduler such as slurm to
send jobs to distributed memory nodes. If used on
a distributed memory cluster then n OPTIM jobs will be run at any given
time, and they will all run on the interactive node in question.
Can be used to run multiple OPTIM jobs on an interactive testing node of a distributed memory
machine.
- CUDA : should be used with keyword SLURM when running GPU jobs on pat.
- CYCLES n: the number of cycles of ncpu or
jpn×nodes OPTIM jobs to run in sampling the stationary point
database.
- CV Tmin Tmax Tinc: calculate the heat capacity in the harmonic
superposition approximation from temperature Tmin to Tmax at
intervals of Tinc.
- CVBREAK: Perform the harmonic heat capacity breakdown assigned to minima
as described in Phys. Rev. E, 95, 030105, 2017.
The temperature for the breakdown is read from Tanal and the sorted minima
fractional contribution to Cv can also be read (default 0.95).
! as in Decoding heat capacity features from the energy landscape.
! DJ Wales Physical review. E (2017) 95, 030105
! (DOI: 10.1103/physreve.95.030105)
- CVMINIMA minstart minend mininc: must be used in conjunction with
CV above.
Calculate the heat capacity as per the CV keyword using sums of local minima
ordered in terms of increasing potential energy.
The first calculation will sum up to minimum minstart, the next
will sum up to minstart + mininc, etc., until we reach
minend.
This keyword provides an automated way to break down the contributions to Cv.
- DB sigmabb: calls
a finite system of dipolar Lennard-Jones dumbbells [#!ChakrabartiFW09!#], where
σ's correspond to the Lennard-Jones parameters.
σaa is set to unity by default.
- DEBUG: turn on debug printing.
- DEGFREEDOMS n: used to specify the number of degrees of freedom, n, in the system. This
should not be used with RIGIDINIT. Instead, the number of degrees of freedom should be specified as an argument
to that keyword.
- DFTBP: Specify that DFTBP is used. PATHSAMPLE will write start.* files for OPTIM rather than appending the coordinates to the odata files. A dftb_in.hsd file must be present.
- DGT DisconnectSources AltPbb Rescale Normalise:
dense optimised graph transformation rate calculation [#!TrygubenkoW06!#].
The four arguments are all logicals, so an example input line might look
DGT T T F F. If true (the default) DisconnectSources specifies that
a full transformation should be performed for each source, disconnecting the
other sources. The resulting rate constants correspond to the KMC result and
kKMC [#!Wales06!#].
If AltPbb is true (default is false) then additional work is done to
maintain precision, which roughly doubles the execution time, but may be needed at low
temperature.
If Rescale is true (default false) an alternative strategy is used
to try and prevent error propagation.
Setting Normalise true (default false) instructs GT2input.f90 to check the normalisation
of branch probabilities. This should not be necessary.
- DIHEDRALMETRIC: use the dihedral angle metric for distances in choosing pairs for connection
in SHORTCUT, UNTRAP, etc.
The metric is the sine of the difference in corresponding dihedral angles divided by two, squared, and
summeed over all dihedrals. If n is also specied then the metric is raised to the power n.
- DIJINITCONT [EXP/n/INDEX/DIHEDRAL n] max p1 p2: continue an initial Dijkstra connection
from information in the existing min.A, min.B, min.data,
ts.data, points.ts and points.min files.
The minimum information required consists of database entries
for the two endpoint minima.
The algorithm used is similar to OPTIM, except that the missing connections can
be sought in parallel OPTIM runs, each of which is itself a separate
Dijkstra connect calculation.
If EXP appears after DIJINIT then the edge weights are based on the
exponential distance; otherwise the distance is raised to the power n, which
defaults to 2; alternatively, INDEX uses
the modulus of the difference between the positions of the minima in the order they were found.
The max parameter (default 100) specifies the number of entries for each
minimum in the sorted list of metric values (default metric is distance).
These values are now dumped in the file pairdist with the identities of the
minima stored in file pairlist.
However, see INITIALDISTANCE for an alternative scheme, which creates the file
allpairs.bin instead of pairdist and pairlist.
This scheme enables much faster restarts if the number of minima grows large before
a connection is found, and limits the memory used to linear scaling rather than quadratic.
If ADDPATH is used on its own before a connection has been
achieved then DIJINITCONT should be specified so that the
pairdist and pairlist files are written.
If the same connection is attempted more than once the same odata file
is used, and the previous calculation is simply duplicated.
The number of parallel searches is equal to the number of processors available.
For the first cycle all the searches would be the same, because there must be a single
gap between the two end minima.
To avoid unnecessary duplication it is best to perform an initial run on
one processor, or a single node, for one cycle (see DIJINITSTART below).
Then restart by simply increasing the number of cycles in the pathdata file
as well as the number of processors.
In the Dijkstra analysis minima with zero connections are removed.
This value can be changed using a DIJKSTRA n line in pathdata.
If p1 and p2 are specified then PATHSAMPLE will calculate the
pairlist and pairdist entries for minima in the range p1 to
p2 (inclusive) write them to files pairlist.p1.p2 and pairdist.p1.p2,
and stop.
The DIHEDRAL argument with optional integer n specifies a metric based on dihedral angles.
The metric is the sine of the difference in corresponding dihedral angles divided by two, squared, and
summeed over all dihedrals. If n is also specied then the metric is raised to the power n.
- DIJBARRIER power:
for Dijkstra shortest path calculations use edge weights corresponding to the
barrier raised to power power. The default for power is 1.0.
Note that this keyword does not run a Dijkstra calculation.
- DIJTS power:
for Dijkstra shortest path calculations use edge weights corresponding to the
energy difference between the next transition state and the global minimum in
the database, raised to power power. The default for power is 1.0,
but large values may be needed to pick out the pathway with the lowest maximum
transition state energy. Using edge weights based on branching probabilities may
not pick out the path with the lowest barrier, because the branching probability can be
significant for minima that only have connections via high energy transition states.
Note that this keyword does not run a Dijkstra calculation.
- DIJCHECKTS n: will check transition states on the Dijkstra fastest path.
Minima with n connections or fewer are ignored, as for the DIJKSTRA keyword.
Designed for use with databases created using DNEBPATH where all the
stationary points are just DNEB images.
- DIJCHECKMIN n: will check minima on the Dijkstra fastest path.
Minima with n connections or fewer are ignored, as for the DIJKSTRA keyword.
Designed for use with databases created using DNEBPATH where the
stationary points may be DNEB images.
- DIJINITCONTFLY exp or n: performs the same function as DIJINITCONT but
calculates distances between local minima on the fly. This approach may be necessary
for initial path runs that produce more that 20,000 minima, where fitting all the
distances into memory becomes an issue.
- DIJINITSTART exp or n max: perform an initial Dijkstra connection run
for the two endpoints specified in odata.start and odata.finish.
In this case the files min.A, min.B, min.data, ts.data (and all
points files) will be created automatically.
Any existing database information will be overwritten.
Note that the odata.start and odata.finish files must contain
the OPTIM keyword DUMPDATA and calculate the normal mode frequencies.
For a CHARMM run the files start.crd and finish.crd are also needed.
The algorithm used is similar to OPTIM, except that the missing connections can
be sought in parallel OPTIM runs, each of which is itself a separate
Dijkstra connect calculation.
If exp appears after DIJINIT then the edge weights are based on the
exponential distance; otherwise the distance is raised to the power n, which
defaults to 2.
The max parameter (default 10) specifies the number of entries for each
minimum in the sorted list of metric values (default metric is distance).
These values are now dumped in the file pairdist with the identities of the
minima stored in file pairlist.
If ADDPATH is used on its own before a connection has been
achieved then DIJINITCONT should be specified so that the
pairdist and pairlist files are written.
This scheme enables much faster restarts if the number of minima grows large before
a connection is found, and limits the memory used to linear scaling rather than quadratic.
If the same connection is attempted more than once the same odata file
is used, and the previous calculation is simply duplicated.
The number of parallel searches is equal to the number of processors available.
For the first cycle all the searches will be the same, because there must be a single
gap between the two end minima.
To avoid unnecessary duplication it is usual to perform the first cycle of an initial
connection attempt on one processor for one cycle.
Then restart using DIJINITCONT
by simply increasing the number of cycles in the pathdata file
as well as the number of processors.
For the first cycle it is also a good idea to use just one double-ended connection
cycle in odata.connect, and increase this value on subsequent cycles
when some intervening minima are known.
Otherwise the first OPTIM job may spend a great deal of time performing
connection attempts that could be spread over multiple processors in a
PATHSAMPLE DIJINITCONT job.
In the Dijkstra analysis minima with zero connections are removed.
This value can be changed using a DIJKSTRA n line in pathdata.
- DIJINITSTARTFLY exp or n: performs the same function as DIJINITSTART but
calculates distances between local minima on the fly. This approach may be necessary
for initial path runs that produce more that 20,000 minima, where fitting all the
distances into memory becomes an issue.
- DIJKSTRA n: perform a Dijkstra analysis to find the path
with the largest contribution to the `SS' rate constant ignoring recrossings. Minima with
n connections or fewer are pruned.
If REGROUPFREE, REGROUPRATE, or REGROUPPE is present
in pathdata then the stationary points will be regrouped first. In this
case the energies in Epath are free energies, and files such as
redopoints and stationary.points are not produced.
Once a path is found the rate corresponding to that path in isolation,
i.e. ignoring other connections, is calculated using the graph transformation
method.[#!TrygubenkoW06!#].
The transition state printed in the fastest path summaries is the one with the lowest energy.
Branching prbabilities between minima are based on all the available transition states that connect
each pair. Hence we include all these transition states when doing a
RETAINONLYSP calculation, which may be based on DIJKSTRA or
KDISTINCTPATHS analysis.
- DIJKSTRALIST n: perform a Dijkstra analysis for all pairs of minima in the file
DijkstraList. Minima with n connections or fewer are pruned.
Minima on the path, the highest energy, and the end minima, are dumped in files
onpath, high, and low. Minima one step away from these sets, two steps, etc.
are also dumped in files with suffices 1, 2, etc.
See also DIJKSTRALIST2.
- DIJKSTRALIST2 n depth: same as DIJKSTRALIST, but the onpath, high, and low
are saved individually for each pair of minima in DijkstraList, instead of dumping them all together.
For example, onpath.0__1.to.7 is the set of minima on the fastest Dijkstra path between minima 1 and 7.
depth is the number of steps away to continue the analysis, maximum 8.
Beware of making excessive files if there are too many entries in the DijkstraList file.
The mean first passage times for the unbranched Dijkstra fastest paths for each pair of minima
are also dumped in Dijkstra.unbranched.MFPT.
The idea is to try and pick out paths that may need protection from renormalisation
in the partial GT procedure, where protection means keeping more neighbouring minima
instead of renormalising them away.
- DIJKSTRAWAIT: perform a Dijkstra analysis to find the path
with the lowest sum of waiting times times initial conditional probability.
- DIJPAIR n1 n2 string n3 : choose pairs of minima for connection attempts based on
the highest barrier on the path that makes the largest contribution
to the `SS' rate constant. The first time a given transition state is the highest,
the connection attempt is between the adjacent minimum.
The next time it is chosen we use the second-nearest neighbours along the path in question.
The parameter n1 determines how many steps away from the transition state we
continue these connection attempts for. The default is n1=1.
Once n1 attempts have been tried for the highest transition state we
move to the second-highest, and so on.
Minima with n2 connections or fewer are iteratively removed before the network
analysis. The default value for n2 is one.
The string argument is optional. If it is the
word `BARRIER' then pairs of minima to connect are sorted according to the
largest barrier
that separates them. Otherwise they are sorted in order of increasing
energy of the minimum that is separated from the product region.
If string is `MAXSEP' then the maximum separation for connection pairs on the
fastest path is read as an integer n3 following this word. The default is to consider all
pairs, but this is probably not appropriate for very long paths.
- DIJPATHPAIRS cut shift barrier: will select pairs of minima for connection
based on the Dijkstra shortest path so long as their distance is greater than
cut and one of the barriers is greater than barrier.
The shift is applied to reduce the corresponding ln rates locally,
or to raise the ts energy if DIJBARRIER is used.
This keyword is designed for use with databases created using DNEBPATH where
we want to try connecting adjacent fictitious minima until the DNEB construction
does not produce any more local minima.
- DIJPRUNE exp or n max p1 p2: using the existing best path calculation
in DIJINITCONT it creates a new file min.retain which only contains minima
on the best path. The weighting option are identical to DIJINITCONT. It requires
that CYCLES is set to 1.
Allows consequently to remove all other minima (useful to keep databases small). Can be used
with PRUNECYCLE and IGNOREPAIRS.
- DIRECTION a: the direction (`AB' or `BA') determines the sense of a path, for
contexts where forward and backward or reactant and product need to be defined. Note the use
of the spectroscopists' convention, whereby `AB' means from B to A . The default is `AB'.
- DNEBPATH: for use with databases using the OPTIM DNEBPATH
keyword. When using SHORTCUT with BARRIER or
MINMAX the DNEBPATH keyword allows the minima directly
connected to the fake transition state to be chosen if they satisfy the other
criteria.
- DOINVERSION: forces inversion check of structures for distance
metric in minpermdist, even for biomolecules.
- DSCALE x: specifies how the separation between local minima, d, is used
to select the pair used in the next attempted connection. If
exp(- (d - x)/x) < a random number drawn from [0, 1] then the distance criterion is accepted.
There is also a criterion for the difference in committor probabilities (see PSCALE).
If d < x then the distance criterion is always satisfied; above x the probability
of acceptance decreases exponentially.
- DUMMYRUN: specifies that no OPTIM jobs should be submitted; the program just
sleeps instead.
- DUMMYTS: specifies that dummy transition state entries should be created for
selected pairs of minima. The corresponding database of minima and dummy transition states
can be refined using any of the usual schemes, e.g. SHORTCUT, FREEPAIRS, etc.
Either BHINTERP or BISECT needs to be specified in the pathdata file,
so that OPTIM jobs only locate new local minima and feed them to PATHSAMPLE
via min.data.info files.
The dummy entries in ts.data include connections for all nearest pairs of minima
for BHINTERP, but only for successive pairs of minima with BISECT.
The dummy energy and log product of mass-weighted Hessian eigenvalues for each
ts.data entry are chosen according to properties of the two minima.
The current energy estimate is the energy of the higher minimum plus the minimum distance
between the two minima.
The estimate for the log product of eigenvalues simply rescales the value for the higher minimum,
effectively removing one degree of freedom.
At some point a decision must be made to start linking the minima via genuine transition
states. To do this we use USEPAIRS, which reads a sequence of stationary points in
the format of the Epath file.
The dummy ts.data and pairs.data files must be renamed or removed.
- DUMPDIHEDRAL: Dump all dihedral angles (not impropers) for minima and transition states in the database
- DUMPGROUPS: specifies that the groups of potential energy minima and transition
states corresponding to groups of free energy minima and transition states should be dumped
to files minima_groups and ts_groups following regrouping.
- DUMPALLPAIRS: will dump the entries in allpairs.bin in plain text format
to enable checks for the INITIALDIST keyword.
- DUMPPB: branching probabilities will be dumped in file Pbranch.temperature.
- EDIFFTOL x: two stationary points will only be identified as the
same structure if their energy difference is less than x and their minimum
distance is less than the threshold specified by GEOMDIFFTOL.
ETOL specifies the same parameter and is provided for backward compatibility.
- ENERGY x: specifies that the microcanonical ensemble at energy x is to be used
to calculate rate constants. Mutually exclusive with TEMPERATURE. See also FREEENERGY.
- ETOL x: energy difference criterion for distinguishing stationary points.
Used with ITOL in PATHSAMPLE.2.0 and GEOMDIFFTOL in
PATHSAMPLE.2.1. EDIFFTOL specifies the same parameter.
- EVCUT x: tolerance for Hessian eigenvalues or frequencies read from
a path.info file by the getallpaths subroutine.
If an eigenvalue is detected that should belong to the positive set by lies below
x (default value
2×10-6) the min-sad-min triple is skipped.
- EVTRUST x: for use with WAITPDF.
Eigenvalues smaller in magnitude than x are considered unreliable and discarded.
An effective eigenvalue and amplitude are added to the first passage time distribution
to correct the mean first passage time (if NGT is run as well) and the normalisation.
- EXEC a: name of the OPTIM executable to be called by PATHSAMPLE.
- EXTRACTMIN n: extract the coordinates of minimum n from
points.min and write them to file extractedmin. If the CHARMM
keyword is present then a pdb format extractedmin.pdb will also be
produced. If n≤ 0 then all the minima present in the points.min file are extracted to extractmin.
The points.min file could still contain extra minima beyond the entries found in min.data which would lead to more extracted minima than expected.
These extra minima won't be used by further PATHSAMPLE or disconnectionDPS jobs and can be ignored or deleted from the end of extractmin.
The numbering of the other minima in extractedmin still corresponds to those found in min.data.
If n = - 123 then a new binary file points.min.new is written from the
entries in the extractedmin file. This setting provides a way to populate
the coordinates for databases created without structures, for example, from a rate matrix.
- EXTRACTMINFILE: extract the coordinates of minima
listed in file extractmin from
points.min and write them to file extractedmin.
- EXTRACTTS n: extract the coordinates of ts n from
points.ts and write them to file extractts.
If the CHARMM
keyword is present then a pdb format extractedts.pdb will also be
produced. If n≤ 0 then all the transition states are extracted to extractts.
If n = - 123 then a new binary file points.ts.new is written from the
entries in the extractedts file. This setting provides a way to populate
the coordinates for databases created without structures, for example, from a rate matrix.
- EXTRACTTSFILE: extract the coordinates of ts
listed in file extractts from
points.ts and write them to file extractedts.
- EXTRAPERM: for use with PERMDIST when allowing the full permutation
group is too permissive. Allows the custom specification of what permutations to
allow. For example, when dealing with rigid bodies, the body might have a rotation.
The permutations are listed in the file extra_perm.allow, which must be present
in the working directory. The first line must specify the number of groups in the
file. This is followed by the specification of the groups, each of which has the
following format:
<Number of permutations in the group><Number of atoms in the group>
<Identity permutation>
<Other permutation>
.
.
.
Each group should probably be a full symmetry group in the mathematical sense, but
this is not checked. For example, if there were two rigid bodies that had the form of pentagonal
bipyramids, with an allowed five fold rotation axis interchanging the radial
atoms, that were 3-7 in each body, extra_perm.allow might be:
2
5 5
3 4 5 6 7
4 5 6 7 3
5 6 7 3 4
6 7 3 4 5
7 3 4 5 6
5 5
10 11 12 13 14
11 12 13 14 10
12 13 14 10 11
13 14 10 11 12
14 10 11 12 13
- FREEENERGY : specifies that the energies in min.data and ts.data
should be treated as free energies for calculating rates and occupation probailities.
The point group orders, vibrational frequency information and moments of inertia are not used
in this case.
Also requires a TEMPERATURE. See also ENERGY for microcanonical ensemble.
- FROMLOWEST (NOLABELS): reads GMIN lowest files and writes PATHSAMPLE points.min
and min.data files. Frequencies are not currently calculated, and so FROMLOWEST
can only be used when NOFRQS is specified. If you GMIN lowest file does NOT contain atom labels
at the start of each line, the NOLABELS option should be used.
- FREEPAIRS x1 x2 x3: connection pairs are chosen based upon the same free energy
regrouping scheme as REGROUPFREE.
x1 corresponds to the free energy barrier height below which free energy
minima are combined.
Free energy minima that lie more than x2 units of energy above the global
free energy minimum group are ignored.
x3 is the energy increment used in the superbasin analysis to estimate
barriers.
Pairs of potential energy minima are chosen from groups based on the ratio
of free energy barrier height to product divided by free energy difference
from product. Hence we highlight groups that correspond to frustration,
with similar energies to the product but high barriers.
Local minima from these free energy groups are then chosen
based upon an additional shortest distance criterion.
Searches are not repeated for the same potential energy minima.
NOTE: for this to work efficiently it is important to have the lowest
minima specified in min.A and DIRECTION set to AB, so that
the A minima are product, and lie at the bottom of the landscape.
See also the PAIRLIST keyword.
- FREEZE n1 n1...: specifies that atoms n1, n2…, should be frozen.
This keyword must come after the NATOMS keyword.
- FRICTION γ: specifies that rate constants should be
calculated using the multi-dimensional version of Kramer's theory described by
Berezhkovskii, Pollak and Zitserman.[#!BerezhkovskiiPZ92!#]
γ is a phenomenological `friction' coefficient in frequency units, which
must correspond to the units for the Hessian eigenvalue product in the
ts.data file. This formulation requires the imaginary normal mode
frequency for each transition state, and the ts.data file must therefore
contain a ninth column corresponding to the unique negative Hessian eigenvalue.
For the CHARMM and AMBER force fields this eigenvalue is the
angular frequency squared in SI units, while for all other potentials it will
be in reduced units. Such databases can be extended without using the `friction'
formulation by setting the IMFRQ keyword (or by using γ = 0).
- GCORES string: needed to specify the number of cores to run on for Gaussian09 jobs
on volkhan (slurm 14) if %NProcshared is greater than one in the gaussian.inp.pre file for
OPTIM. To use 8 cores on an exclusive node set the string to ' -c8 ' in pathdata
and in the slurm batch script use:
#SBATCH -N<number of nodes>
#SBATCH –tasks-per-node 1
#SBATCH –exclusive
This option also works for the interface to the xtb program with OPTIM.
- GEOMDIFFTOL x: two stationary points will only be identified as the
same structure if their minimum separation is less than x and their energy
difference is less than the threshold specified by EDIFFTOL.
- GETMINFRQS : used with CHECKMIN. For all checked minima the frqs.dump file
is created and then all frequencies are written to a new file frqs.min.
Every frequency is paired with the minimum id, and it can then be used to create a min.data file
with frequencies, if OPTIM and PATHSAMPLE were run with NOFRQS.
Frequencies are only written, if the CHECKMIN routine converged. The odata.checksp file
needs to contain DUMPFRQS.
- GETTSFRQS : used with CHECKTS. For all checked minima the frqs.dump file
is created and then all frequencies are written to a new file frqs.ts.
Every frequency is paired with the minimum id, and it can then be used to create a ts.data file
with frequencies, if OPTIM and PATHSAMPLE were run with NOFRQS.
Frequencies are only written, if the CHECKTS routine converged. The odata.checksp file
needs to contain DUMPFRQS.
- GT n1 n2: specifies that the graph transformation approach[#!TrygubenkoW06!#]
should be used to calculate rate constants.
Minima with n1 connections or fewer are iteratively removed before the rate
calculation. n2 specifies that the rates should be calculated every n2 cycles
during the refinement of the stationary point database. For large databases making n2 equal to one
could slow the sampling down significantly.
To run a GT calculation without changing the stationary point database set CYCLES 0.
See keyword GT2 for a more general implementation of graph transformation.
The rates produced from the GT keyword correspond approximately to the
kNSS rate constants [#!TrygubenkoW06!#,#!Wales06!#].
Branching probabilities for return to the same initial state are set to zero,
and all the other connections are renormalised accordingly. Hence the final branching
probabilities for a given source include arbitrary revisits to that source before
a path reaches a different source or a product, so they do not quite correspond to
committor probabilities. In practice, the resulting rate constants generally seem to
be close to the
kNSS values.
- GT2 This keyword has been removed. Please use SGT,
DGT or SDGT.
- GT2RSwitch x : specifies that the generalised graph transformation
routines switch from single to double precision when the density reaches x.
The default value is x = 0.08.
See keyword SDGT and subroutine GT2 for more information.
- GT2PTOL x: specifies the threshold for deciding that a node is dead
in the generalised graph transformation routines. The default value is x = 0.00001.
- IGNOREPAIRS value : value is either TRUE or FALSE. If set to TRUE, previously
searched pairs will be ignored in the Dijkstra analysis in the Dijinit subroutine.
- IMFRQ: specifies that the ts.data file should contain a ninth
column containing the unique negative Hessian eigenvalue. This
is the eigenvalue corresponding to the imaginary normal mode frequency.
See also the FRICTION keyword, which implies the same format for
ts.data and calculates the rate constants using a phenomenological
description of dynamical solvent friction.
- INITIALDISTANCE disbound: use a fictitious fixed distance disbound for pairs of minima
in DIJINITSTART and DIJINITCONT runs.
The true distance is only calculated when the connection appears in the Dijkstra missing connection
best path, and the Dijkstra analysis is repeated until all the steps in the best path involve connected minima
or pairs where the true distance is known.
With a small disbound we calculate all the true distances, because the small distance makes it into the best path.
With a large disbound we will accept larger true distances into the best path, so fewer distance calculations will be needed.
The objective is to stop pair distance calculations from slowing down initial pathway attempts,
where a large number of permutable groups can be a problem, especially for LPERMDIST.
The current distance values are saved in file allpairs and held in memory in vector ALLPAIRS.
If a database grows very large without an initial connection being found, this vector may cease to
fit in available memory.
The algorithm is similar to the one presented by Noe et al. JCTC, 2, 840, 2006.
If disbound is negative then all distances are calculated.
- INTCONSTRAINT intconstrainttol intconstraintdel intconstraintrep intconstrainrepcut
intconsep intrepsep
specifies quasi-continuous interpolation via an auxiliary constraint potential, as
for OPTIM. Note that some of the OPTIM parameters are not needed
for PATHSAMPLE, and are omitted.
In PATHSAMPLE this keyword is only used to define the QCI metric, which can
be used in selecting minima for connection. It is never used with any intervening
images. PATHSAMPLE does not use congeom or congeom.dat files:
it simply calculated the QCI metric based on pairs of local minima.
intconstrainttol is used to determine constrained distances. The deviation
of the distance between a given pair of atoms
in all reference structures from the average must be less than
intconstrainttol, default 0.1, for this separation to constitute a constraint.
If a percolating network of constraints does not result then intconstrainttol
is increased by 10% and the analysis repeated.
intconstraintdel multiplies the constraint penalty term in the auxiliary potential,
default 10.0.
intconstraintrep multiplies the repulsive penalty term in the auxiliary potential,
default 100.0.
intconstrainrepcut is the cutoff for the repulsive penalty term in the auxiliary potential,
default 1.7.
intconsep is the maximum difference between the order number of atoms for
which constraints are allowed. The default is 15, which seems appropriate for
biomolecules. However, if a congeom file of reference minima is specified, or
a corresponding congeom.dat file is present, then intconsep can be set
greater than the total number of atoms.
intrepsep is the minimum difference in order number of atoms for which repulsive
terms are added to the potential. The default is zero; there are no repulsive terms
between constrained atoms.
- ITOL x: principal moment of inertia tensor difference criterion for
distinguishing stationary points. All three values are compared for stationary points
in the initial setup phase. However, unlike PATHSAMPLE.2.0, the alternative
criterion specified by GEOMDIFFTOL is used to distinguish stationary points
in PATHSAMPLE.2.1.
- KDISTINCTPATHS npaths mode: specifies the use of the "k distinct paths" shortest paths algorithm to
find the dominant A<-B paths in a transition network, where each path is distinguished on the basis of
having a different rate-limiting step. npaths is the maximum number of shortest paths to be computed - note
that the algorithm will terminate automatically when a cut is induced in the network by the iterative blocking of
transition states. Also note that this may happen after very few iterations if the network is only sparsely connected
in the dynamical bottleneck region, and this event strongly indicates that further sampling of the stationary point database is
required to obtain a sensible representation of the dynamics. mode determines the definition of what is considered
a "rate-limiting edge" (RLE); 0: highest-energy TS is the RLE; 1: highest energy barrier defines the RLE; 2: edge with largest
weight defines the RLE.
Produces file "Epath.xxxx", where xxxx is the path number (iteration), in the usual Epath format.
Also produces output file "rate_lim_cut.dat" with format:
ID of rate-limiting TS / weight of rate-limiting edge / total weight of path / iteration number.
Use in conjunction with DIJKSTRA 0. Coded by Daniel.
- KMC n1 x n2: perform n1 KMC runs having iteratively removed
minima with n2 connections or fewer.
x specifies a pair threshold for the product, p, of forward and backward
branching probabilities between two minima.
If p is greater than x then a leapfrog move will be attempted to a
different connected minimum when either of the pair is encountered.
Setting p to one or more means that leapfrog moves will not be used.
No additional stationary points will be calculated in a KMC
run: the CYCLES keyword is ignored if it is present in odata.
Only one of the GT, GT2, KMC and KMCcommit keywords should be
present in a given pathdata file.
- KMCcommit n1 x1 x2 x3 x4 n2: specifies a rate constant calculation using
committor probabilities and appropriate waiting times. This subroutine can calculate
both `SS' and `NSS'-type rate constants.[#!TrygubenkoW06!#]
n1 is the number of KMC trajectories to run. If n1 is zero then
the waiting time in a given minimum is the the escape time
to its connected neighbours. Combined with the committor probabilities,
these waiting times give `SS' rate constants. If n1 is greater than zero then
the waiting times are calculated for KMC runs that terminate as soon as they reach
a minimum in either the A or the B region. The resulting rate constants
are described as `NSS'.[#!TrygubenkoW06!#]
x1 is the maximum number of iterations allowed in the committor probability
calculation; it is a real number because the value required may exceed the size allowed
for an integer.
x2 is a convergence condition for the committor probabilities, and is defined
in terms of the percentage change between iterations.
x3 is the ω parameter used in the successive overrelaxation calculation
of the committor probabilities. It should be greater than or equal to one, but less than 2.
The default value is one, which results in Gauss-Seidel iterations.
x4 specifies a pair threshold for the product, p, of forward and backward
branching probabilities between two minima.
If p is greater than x then a leapfrog move will be attempted to a
different connected minimum when either of the pair is encountered.
Setting p to one or more means that leapfrog moves will not be used.
n2 determines the minimum connectivity allowed for local minima to be
admitted into the calculation. The minima are iteratively pruned to remove those
with n2 connections or fewer.
No additional stationary points will be calculated in a KMCcommit
run: the CYCLES keyword is ignored if it is present in odata.
Only one of the GT, GT2, KMC and KMCcommit keywords should be
present in a given pathdata file.
- KSHORTESTPATHS npaths nconnmin extraprinting: calculate the npaths paths
with the largest contributions to the rate constant when intervening minima are
put in steady state. Coded by Dr Joanne Carr.
nconnmin specifies that minima with nconnmin connections or fewer
should be disregarded in this analysis. Setting the optional argument extraprinting to "T"
turns on extra output printing for each path (off by default).
- LOCALPERMDIST thresh cutoff: minimise distances between end points
prior to DNEB interpolation using local alignment.
thresh is used to define the tolerance for a `perfect' alignment
in routine minpermdist, which is called for subsets of atoms
rather than all at the same time for PERMDIST.
The threshold used is actually the number of atoms in the permutable
group in question times thresh.
The routine minpermrb cycles through the permutable groups,
calling minpermdist for each one, augmented by any other permutable
groups that overlap, as for any groups with additional pair swaps, such
as valine and arginine.
Atoms that are equidistant from the permutable atoms in the two end points
are also added to the set if the distance lies within a cutoff
radius cutoff.
They are added in increasing distance order, and removed one at a time
if the resulting subsets of atoms cannot be aligned `perfectly' between
the two endpoints.
Using cutoffs of 2.5 and 5 Å gives different internal rotations for
an arginine group, but with similar barriers.
The default values are 0.1 and 5.0 for thresh and cutoff,
respectively.
Requires the auxiliary file perm.allow to specify permutable atoms, otherwise
all atoms are assumed to be permutable. The absence of a perm.allow
file is considered a mistake for CHARMM runs.
See also LPERMDIST, which effectively replaces LOCALPERMDIST.
- LOWESTFRQ: specifies that the lowest positive eigenvalues associated
with the Hessian and mass-weighted Hessian should be read from min.data.info
files and recorded in min.data.
These eigenvalues can then be used in combination with DUMMYTS
to estimate the energy and frequency factor associated with dummy transition states.
The LOWESTFRQ keyword must also appear in odata.bhinterp or odata.bisect.
The current estimate for dummy transition state parameters does not require these
extra data.
- LPERMDIST n thresh cut orbittol: provides the preferred local
permutational alignment procedure.
PERMDIST may be a better option for large biomolecules until the
database actually contains a connection (see end of paragraph).
n is the maximum number of extra atoms to include in running
minperm for each permutable group (default 11, a value that may be needed to treat phenylalanine), rather than
treating the whole system at the same time, as for PERMDIST.
The extra atoms that define the neighbourhood of the permutable group are
added one at a time in order of their distance from the centre of coordinates of the
group, averaged over the start and finish geometries.
If the optimal aligned distance is greater than thresh (default value 0.2)
then the atom is not included in the neighbour list.
This procedure continues until there are n atoms in the neighbour list,
or no more atoms within the cutoff distance cut, default 10.0.
The parameter orbittol is the distance tolerance for assigning atoms to orbits
in the myorient standard orientation routine, default value 0.001.
This keyword requires the auxiliary file perm.allow to specify permutable atoms, otherwise
all atoms are assumed to be permutable. The absence of a perm.allow
file is considered a mistake for CHARMM runs.
WARNING: LPERMDIST can slow down DIJINITCONT runs dramatically
for large biomolecules with many permutable groups if the database grows beyond a
thousand minima without finding an initial connection.
In fact, PERMDIST in the pathdata file may be OK (and much faster)
so long as LPERMDIST is used for OPTIM, which would fix any
bad permutational alignments before starting a transition state search.
- MACHINE: if specified, certain files are written and read as
binary rather than plain text.
- MACROCYCLE n: sets up permutational isomers for a
macrocyclic system. The number of repeat units in the macrocycle is given by
n (e.g. n=4 for cyclo-[Gly4]). The order of the atoms in
each repeat unit must be the same and these permutations should not be added to
the perm.allow file. This keyword has only been tested on cyclic peptides using
the AMBER potential.
- MAKEPAIRS pairfile: use in conjunction with ADDMIN or READMIN
to produce an Epath-style pairfile which can then be used with USEPAIRS
to connect a chain of minima together. The minima from the min.data.info are
read into the PATHSAMPLE database as normal, but in addition the pairfile
records the indices of the minima in the order in which they appeared in
min.data.info. Running PATHSAMPLE with USEPAIRS then connects every pair of
minima from the original min.data.info file, in the order they appeared. Minima
may appear multiple times in the same pairfile if they appear multiple times in
min.data.info. The exception is consecutive duplicate minima, which are
omitted.
- MAKESTART : put the coordinates for the initial minimum in a start file
rather than at the end the odata file. For some potentials OPTIM reads from
the end of odata; for others it is more convenient to use a start file,
and for the OPTIM potentials in MANYBODY and MANYBODYSTART you
can use either.
- MAXBARRIER x: use in the post-processing analyses to set a criterion for excluding transition
states if the barriers from both connected minima exceed x. The threshold x is reset to infinity
on converting potential energies to free energies during a run.
- MAXDOWNBARRIER x: specifies a maximum downhill barrier
of x for DIJKSTRA analysis.
The downhill barriers for the path with the largest contribution to the
non-recrossing steady-state rate constant are sorted and those that exceed
x are analysed further. The transition state in question is removed
and the DIJKSTRA analysis performed again if a connected path is still possible.
If a connected path is no longer possible the transition state in question is
reinstated. A list of `shiftable' transition states is provided at the end.
Note that the order in which transition states are removed could affect this list,
so that other combinations of `shiftable' transition states may be possible.
The downhill direction can only be defined locally once the complete
DIJKSTRA path is known, since `downhill' is determined by the energies
of the two endpoints.
- MAXTSENERGY x: specifies that transition states above energy x should
not be included in the database. Probably most useful with CHARMM, where genuine
transition states with silly energies can certainly occur and give rise to underflow.
This keyword performs the same function as TSTHRESH, and is provided because
OPTIM uses MAXTSENERGY.
- MERGEDB dir: merge the min.data, min.ts, points.min,
points.ts, min.A and min.B files from directory dir with
those in the current directory. Common stationary points are identified and
not repeated. Merged results are created corresponding to all six of the above files.
This keyword can be used to remove duplicate stationary points from a database if
the allowed tolerances or permutations change.
In this case you should start with a completely empty database, and merge in the original
database in question, which will check for duplicates as it proceeds.
- MERGEDBLIST dir: merge selected min.data, min.ts, points.min,
points.ts, min.A and min.B files from directory dir with
those in the current directory. Common stationary points are identified and
not repeated. Merged results are created corresponding to all six of the above files as for
MERGEDB, except that only stationary points specified in files min.merge and
ts.merge are included. Specifying a transition state without one or both the corresponding
minima is allowed, and will generate a WARNING. The merge files must have the number of entries
on the first line and then the indices of the minima and transition states to be merged follow,
one per line, in any order.
- MFPTLIST: use NGT for all pairs of minima specified in file MFPTlist
(note that the number of NGT calls is quadratic in the number of minima specified).
See also MFPTLISTA, MFPTLISTB, ALLMFPT.
- MFPTLISTA: use NGT for all minima specified in file MFPTlist to the A set.
See also MFPTLIST, MFPTLISTB, ALLMFPT.
- MFPTLISTB: use NGT for all minima specified in file MFPTlist to the B set.
See also MFPTLIST, MFPTLISTA, ALLMFPT.
- ALLMFPT: use NGT for all distinct pairs of minima in min.data.
- MINBARRIER x: use in the post-processing analyses to set a criterion for excluding a transition
state if the barrier in either direction is lower than x. Designed to allow exclusion of TSs
present in the database that lie lower in potential energy than the connected minima, by setting x to zero.
The default value is (basically) minus infinity. Note that a transition state may legitimately lie lower
than the connected minima in free energy.
- MINGAP x RATIO: sets a cut off for what connections should be tried in a Dijinit run.
Either x is the minimum distance between two minima for a connection run to be started, or if RATIO
is used, x is the fraction of the maximum pair distance that is then used as minimum. Should be used to
focus searches on large gaps in the database and ignore small connections entirely.
- MLPB3 specifies a multi-layer perceptron neural network
with in input nodes, hidden hidden nodes, out output nodes, regularisation constant
λ and bias nodes for the hidden and output sums.
The number of variables is hidden×(in+out)+1,
and PATHSAMPLE needs this keyword to deal with input/output
for the OPTIM VARIABLES format. There is another keyword
MLP3, which is exactly the same without the bias nodes.
- NATOMS n: the number of atoms in the system; must be present.
- NATOMS_CHAIN n: the number of atoms in the original protein chain if the system contains
both protein and non-protein (e.g. cofactor) atoms. To be used in conjunction with CHECKSP_MUT.
- NATOMS_NEW n: the number of atoms in the system after a mutation. To be used in conjunction
with CHECKSP_MUT.
- NCONNMIN n: specifies that minima with n connections or fewer
should be disregarded in regrouping and rate calculations. This parameter
can be set via other keywords, but NCONNMIN is provided for use with
SGT, DGT, SDGT and also PFOLD. The connection condition is evaluated recursively.
- NEWCONNECTIONS n maxattempts firstmin:
this keyword provides a fully parallelised way to grow a database using single-ended
transition state searches.
It therefore provides an alternative to procedures such as SHORTCUT and
UNTRAP.
Starting from firstmin (default 1) PATHSAMPLE will step through all the
known minima and perform up to maxattempts transition state and pathway
searches (default 10) according to the template odata.tspath file, which must be
present in the working directory.
The next minimum will be considered if the current minimum has
n or more connections (default 0).
for which the default is 10.
The odata.tspath file needs to contain OPTIM PATH
and DUMPALLPATHS directives
so that a path.info file is created.
- NGT nconnmin disconnectsources density size compressedrowswitch: specifies that the new
graph transformation approach should be used to calculate rate constants.
Minima with nconnmin connections or fewer are iteratively removed before the rate calculation.
This calculation produces
kSS,
kNSS and
kKMC for both
A←B and
B←A, as well as committor probabilities.
If disconnectsources is true (default is false) then
kKMC is also
calculated. For single sources
kKMC is the same as
kNSS.
The algorithm switches from sparse to dense storage when the number of connections
exceeds density times the maximum possible value, with the restriction that
there must be size minima or fewer remaining. The default values for density
and size are 0.3 and 11000. Within the initial sparse storage scheme, the algorithm will use
compressed row (vector) storage instead of full, rectangular matrix storage if the memory required for the
two rectangular matrices at the start of the NGT calculation would exceed compressedrowswitch Gb.
See also keyword GT2 for the original graph transformation calculation of
kKMC.
The committor probabilities for the end point minima, which are obtained on removing all the
intervening minima, are written to two files: commit.ngt.AB for direction B to A and commit.ngt.BA for the opposite
direction. The entries for the intervening minima are zeroes. These files have the same form and ordering as commit.data.
The PFOLD keyword (see below) may be given in addition to NGT, in which case a commmittor probability
calculation using the specified control parameters will immediately follow the NGT calculation, and
will be seeded with the known (and fixed) committor probabilities from NGT for the end point minima.
A file commit.data, with one entry for each minimum in the pre-resorting order, will be written at the
end of this calculation containing the committor probabilities for the direction specified via DIRECTION.
- NGTREMOVELIST: will run NGT to disconnect the minima listed (one per line) in the file
NGTremovelist, which must be in the working directory. The remaining non-zero branching probabilities
are dumped in file Pbranch.removedlist.map, with the numbering scheme corresponding to the original
min.data file.
- NGTKEEPLIST: will run NGT to disconnect the minima not listed (one per line) in the file
NGTkeeplist. This keyword performs partial graph transformation and dumps new database
min.data.new, ts.data.new files, which contain free energies that reproduce the
state to state rates defined by renormalised branching probabilities and waiting times from GT,
along with the free energies for the renormalised minima that reproduce the stationary distribution.
See also DIJKSTRALIST for a keyword to suggest entries for the NGTkeeplist file.
- NGTSTEPS nconnmin disconnectsources density size compressedrowswitch: specifies that the new
graph transformation approach should be used to calculate the kinetic activity (mean number of steps).
The options are the same as for NGT above. We simply set all the waiting times to unity, so that
we obtained the number of steps multiplied by the path probability, averaged over all paths.
This result is also equivalent to a discrete time Markov chain with a unit lag time.
- NGTEDGES nconnmin disconnectsources density size compressedrowswitch: specifies that the new
graph transformation approach should be used to calculate the mean first passage path action.
The options are the same as for NGT above.
This formulation gives the sum of edge weights (rewards) multiplied by the path probability, averaged over all paths,
the mean first passage reward (MFPR).
To obtain the mean first passage path action the edge weight is
-ln Pαβ,
for the branching probability
Pαβ. The MFPT values printed are actually MFPR,
and the `rates' are reciprocals. The last two columns in the NGT.rates.new file
obtained with RATESCYCLE become
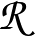
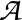
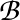 and
and
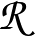
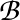
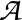 , respectively.
, respectively.
- NINTS n: the number of internal coordinates.
- NIMET NIHEAM7 : these keywords ensure that the EAM4 and EAM7 potentials for hydrogen on a nickel surface
(see OPTIM manual for more details) get the right atom symbols when called from pathsample.
- NOFRQS: specifies that frequencies are not present in path.info files.
Used for very large systems where frequency calculations are not feasible in conjunction
with the same OPTIM keyword.
The log product of positive eigenvalues is set to unity where it is read in from
ts.data and min.data in setup and Dijsktra for consistency.
- NORGFLIST file: the minima listed in file (one per line) will not be
allowed to regroup during the REGROUPFREE procedure.
- NOINITIALDISTPERMNDG: turns off permutational alignment for
calculation of distances with the INITIALDISTANCE keyword coupled to
DIJINITCONT, except for stationary points that are close enough in energy to
be possible permutation-inversion isomers. Hence we should not create duplicate entries
for stationary points in the database, but otherwise the cost of permutational alignment
is saved, which is very important for large molecules and lengthy initial pathways.
- NOINVERSION: turns off inversion of structures for distance
metric in minpermdist.
- NOTRANSROT: forbid overall translation/rotation in alignment and distance
calculations.
- NTIP 4: calls the TIP4P model of water molecules.
- OHCELL : allow point group operations for a cubic
supercell in subroutine minpermdist.f90 permutational distance minimisation.
- OPEP : required for OPEP and HiRE-RNA. Without this keyword PATHSAMPLE
will not create the required start file for OPTIM.
- ORBITGEOMTOL x: sets the cutoff value for assigning atoms to the
same orbit when analysing the standard orientation in routine myorient
when LPERMDIST is specified.
The default value for x is 0.3.
If x is too small it is possible for permutational isomers to be missed.
- ORDER x: reads in an order parameter threshold. Currently not used.
- PAHA n: calls a finite system of polycyclic aromatic hydrocarbons (PAH) interacting
via a general anisotropic potential developed from first principles [#!TottonMK10!#]. The PAH ID
n defines the PAH molecule: 1 for benzene is the only option currently available.
- PAIRLIST n: specifies that the list of minima for subsequent connection attempts
should be regenerated every n cycles. The default is now to update the list every cycle.
Use n=0 for the list to remain
fixed throughout the run unless we run out of candidates. The PAIRLIST option has been
implemented for the FREEPAIRS, SHORTCUT, UNTRAP and
DIJPAIR keywords and for the default connection
strategy based on differences in the committor probability. It has not been
implemented for the CONNECTREGION keyword.
- PAP npatch alpha: specifies a patch anti-patch potential.
Each body consists of a Lennard-Jones core, range parameter alpha,
with npatch patches and npatch anti-patches..
- PBS jpn: specifies the number of jobs to run on each core for a
calculation using a scheduler such as PBS to run OPTIM jobs on
distributed nodes. The cores
must be defined by the job submission script in the nodes.info file.
Unless you really want to run more than one job per available core at a time then
jpn can be omitted: the default value is one. PBS has been replaced by slurm on
most clusters.
- PERMDIST x: specifies that all distance metrics should include minimisation
with respect to permutational isomerisation, as well as optimal orientation.
Should be used with the same keyword in OPTIM.
Requires the auxiliary file perm.allow to specify permutable atoms.
The first line of the perm.allow file must contain an integer
that specifies the number of primary groups of interchangeable atoms.
The groups then follow, each one introduced by a line with two integers p and s
that specify the number of permutable atoms in the primary group and the number of other sets
of permutable atoms associated with the primary set.
s may be zero.
Each secondary set of permutable atoms has p members.
The following line contains the indices of the p permutable atoms
in the primary set and then
the indices of the atoms in each of the s secondary sets, one set at
a time.
The parameter x is the distance tolerance for assigning atoms to orbits
in the myorient standard orientation routine.
If x is too small it is possible for permutational isomers to be missed.
For the phenylalanine example illustrated above we must allow three other pairs of
atoms to exchange if we swap 7 and 8. Hence a suitable perm.allow entry is
1
2 3
7 8 5 6 1 2 3 4
Here n = 2 and s = 3: if we exchange 7 and 8 then we must also exchange 5 and 6,
1 and 2, and 3 and 4. There are two atoms in each of the three secondary sets,
since we have specified 7 and 8 as the two primary atoms.
Here is an example perm.allow file for a water trimer using
the flexible QTIP4PF potential, where the energy is invariant to permutations
of water molecules and to exchanges of hydrogens in the same molecule. However,
hydrogens cannot exchange between different oxygens:
4
3 2
1 4 7 2 3 5 6 8 9
2 0
2 3
2 0
5 6
2 0
8 9
The first group of three oxygens has two atoms that must move with each oxygen,
i.e. atoms 2 and 3 for oxygen 1, etc. Hydrogen permutations for each oxygen are
allowed by the three following groups. This scheme allows atoms to appear in more
than one group. There must be a group containing each complete set of permutations
in order for permutation-inversion isomers to be recognised. The format
is compatible with an older scheme, where only pair swaps were allowed for
associated atoms, but now allows for more general permutations.
Scripts to generate allowed permutations automatically for CHARMM and AMBER are available from
the group web site. It is essential to use symmetrised versions of the corresponding
force fields!
- PERMISOMER: equivalent to PERMDIST above, expect that
distance minimisation
with respect to permutational isomerisation is only carried out for stationary
points with energies that differ by less than the parameter specified by EDIFFTOL.
Hence permutational isomers should be identified, but the distance between minima with
different energies is not minimised with respect to atom permutations.
This option is provided for cases where the number of permutations is too
minimise distances with respect to permutations in general.
Should be used with the PERMDIST keyword in OPTIM.
The auxiliary file perm.allow specifies which atoms may be permuted; see
PERMDIST above.
Note that the NATOMS keyword must precede PERMISOMER in the
pathdata file.
- PERSIST einc pethresh peqthresh perthresh :
Persistence analysis along the lines of Cazals et al.
We perform a persistence analysis using superbasins at energy increments of einc
up to a maximum energy range of pethresh.
This analysis uses the persistence for each minimum, defined as the barrier to the transition state
where that minimum can access a lower-energy minimum.
Output files are min.persist, min.persist.select and min.include. The latter two
include only some of the most persistent minima, selected based on a lower bound
equilibrium occupation probability peqthresh and a lower bound, perthresh, on the barrier
height defined by the persistence (actually the deviation from the diagonal in a
persistence diagram, which is the same aside from a factor of
 ). The usual criteria for transition
states and the connectivity of minima also apply.
The file min.include has a format suitable for reading via the
PICK and IDMINFILE keywords in disconnectionDPS. The format of the min.persist* files
is: rank (ordered by persistence), energy, energy+barrier, barrier(=persistence), index of the minimum,
deviation from the diagonal. See also PERSISTEXACT
). The usual criteria for transition
states and the connectivity of minima also apply.
The file min.include has a format suitable for reading via the
PICK and IDMINFILE keywords in disconnectionDPS. The format of the min.persist* files
is: rank (ordered by persistence), energy, energy+barrier, barrier(=persistence), index of the minimum,
deviation from the diagonal. See also PERSISTEXACT
- PERSISTEXACT ALL:
For use with PERSIST; specifies that the barrier heights used to define the persistences
should be obtained via an efficient exact method, rather than the approximate method using
a superbasin analysis. Note that the values of einc and pethresh from PERSIST are not
used here, but must still be present if you want to specify values for peqthresh and perthresh.
The optional argument ALL instructs the program to report persistences for
all the valid minima in the network; the default is to report values only for the minima in the same connected
component as the global minimum (which matches the situation for the approximate method).
- PERTURB x: initial maximum perturbation of the Cartesian coordinates of a minimum,
when looking for a new connected minimum via a single-ended transition state search.
- PFOLD n1 n2 x PFOLDCONV: provides control parameters for the calculation
of committor probabilities for local minima. The calculation
may be performed on-the-fly during the construction of a stationary point database (if used together
with CYCLES <non-zero value>) or as a post-processing analysis (CYCLES 0).
All new local minima are assigned an initial value of zero.
If a file commit.data exists in the working directory then initial values at the start of the run
will be taken from there.
n1 iterations are performed every n2 cycles with parameter ω in
the successive overrelaxation equal to x.
PFOLDCONV is the threshold on the largest fractional difference between Pfold values at
consecutive iterations for early convergence of the algorithm (before the full n1 iterations
have been performed). The default value is 0.0001.
Note that a valid solution also exists for all committor probabilities equal to unity.
I have seen the iteration converge to this solution for a very small database that
has just been created with DIJINIT.
A file commit.data, with one entry for each minimum under consideration in the pre-resorting order,
will be written at the end of a
committor probability calculation for the pathway direction specified in DIRECTION.
The PFOLD keyword may be given in addition to NGT, in which case a commmittor probability
calculation using the specified control parameters will immediately follow the NGT calculation, and
will be seeded with the known (and fixed) committor probabilities from NGT for the end point minima.
- PLANCK h: specifies the value of Planck's constant in units of the
prevailing energy unit times seconds. This numerical value is required for
regrouping free energies. For example, for the CHARMM potential h needs
to be
9.546×10-14 s kcal/mol
See §
![[*]](crossref.png) for more details.
for more details.
- PRINTSUMMARY: prints (to standard out) some information about the kinetic transition
network, including the number of minima and transition states satisfying the various thresholds, the highest
and lowest TSs, and some information about the connected components present. The analysis is performed
after the SETUP routine has been passed through.
- PRUNECYCLE n: repeats the Dijkstra analysis for DIJPRUNE n times and records all minima, on the so found n
fastest paths (all missing connections can be used once overall). Default is 5 cycles.
- PULL : add this keyword if the system has a non-zero static force
added to the potential. PATHSAMPLE.2.1 only uses this keyword to determine the
number of zero Hessian eigenvalues (four). The attached atoms and force magnitude
are not required.
- PSCALE x: determines how the difference in committor probabilities,
p, is used to select the pair of minima used in the next attempted connection. If
p/x > a random number drawn from [0, 1] then the distance criterion is accepted.
There is also a criterion for the minimum distance (see DSCALE).
If p > x then the committor probability criterion is always satisfied; below x the probability
of acceptance decreases linearly.
- QCHEMSCF qchemjob sys nao nmo nalpha nbeta:
specifies the electronic structure landscape defined by QCHEM for the SCF solution landscape.
qchemjob is a string specifying how to run
the QCHEM executable, as for conventional geometry optimisation,
and sys is a string specifying the QCHEM input file.
These strings are not used by PATHSAMPLE, they just enable us to duplicate the corresponding line from
the odata file.
nao nmo nalpha nbeta are the number of atomic orbitals, molecular orbitals,
alpha spin, and beta spin electrons, respectively.
These values determine the number of molecular orbital coefficients, which are saved as references in
points.min and points.ts,
and the actual number of variables on the constraint surface.
The initial molecular orbital coefficients are communicated to OPTIM in start files.
The file qcoverlap is needed in the current working directory for
PATHSAMPLE to calculate distance metrics between solutions.
These metrics are specified by the keyword:
- QCMODE nmode
where values of nmode 2 and 3 specify metrics based on wavefunction and density distance, respectively.
Note that QCHEM uses a scratch file called $QCSCRATCH/qchemjob.
Multiple QCHEM jobs with the same job name will therefore fail.
For standard PATHSAMPLE runs with SLURM a unique string is appended to the input files, so
multiple QCHEM jobs will work. However, only single jobs can work with RUNLOCAL.
- QCHEMGHF qchemjob sys nao nmo nelec:
specifies the electronic structure landscape defined by QCHEM for the SCF GHF solution landscape.
qchemjob is a string specifying how to run
the QCHEM executable, as for conventional geometry optimisation,
and sys is a string specifying the QCHEM input file.
These strings are not used by PATHSAMPLE, they just enable us to duplicate the corresponding line from
the odata file.
nao nmo nalpha nbeta are the number of atomic orbitals, molecular orbitals,
and number of electrons, respectively.
These values determine the number of molecular orbital coefficients, which are saved as references in
points.min and points.ts,
and the actual number of variables on the constraint surface.
The initial molecular orbital coefficients are communicated to OPTIM in start files.
The file qcoverlap is needed in the current working directory for
PATHSAMPLE to calculate distance metrics between solutions.
These metrics are specified by the keyword:
- QCMODE nmode
where values of nmode 2 and 3 specify metrics based on wavefunction and density distance, respectively.
Note that QCHEM uses a scratch file called $QCSCRATCH/qchemjob.
Multiple QCHEM jobs with the same job name will therefore fail.
For standard PATHSAMPLE runs with SLURM a unique string is appended to the input files, so
multiple QCHEM jobs will work. However, only single jobs can work with RUNLOCAL.
- RANDOMMETRIC n: for DIJINITCONT jobs this keyword limits
the number of metric calculations for each new minimum to n. The n
other minima are chosen randomly. The cost of adding each new minimum to the
database is therefore constant, rather than growing linearly with the number
of minima in the database. If n is less than the max
parameter for DIJINITCONT then the metric values are padded with
enormous values.
- RANROT n: for use with PERMDIST. Specifies
that n random orientations will be tried as well as the
initial standard alignment in minpermdist.f90. Several hundred
random orientations may be needed to find the best translational/rotational/permutational
alignment of end points.
- RATESCYCLE t1 t2 ... tn : calculate rate constants
at the end of every cycle for temperatures t1, t2, etc.
Each temperature and forward and backward rate constants are dumped to
file NGT.rates.new.
Any number of temperatures can be specified, so long as continuation
lines are used if the line length exceeds 80 characters.
This keyword was introduced to make the LJ38 demonstration cleaner.
- RBAA: specifies the number of rigid bodies being considered in the angle-axis. NB: this is not part of the GENRIGID framework.
- RBSYM: specifies that internal symmetry operations permuting equivalent sites for
rigid-body building blocks exist. The operations are read in from a file rbsymops, which must
exist in the working directory. The first line of the file is an integer that specifies the number
of operations generated by proper axes of rotation including the identity operation; the operations
are then read in one at a time in a representation consisting of a unit vector, defining the
axis in the reference frame, and an angle in degrees, describing the magnitude of rotation about
that axis. The first operation has to correspond to the identity operation.
This keyword has to be combined with PERMDIST so that structures are aligned and
a distance metric considered based on permutations of identical rigid
bodies and any internal symmetry operation of each rigid body that is a symmetry
of the potential energy function. NB: this is not part of the GENRIGID framework.
- READMIN file: reads in the data for one or more minima from file
in OPTIM min.data.info format and then stops.
Any number of min.data.info files can be concatenated into file.
New entries will be created in min.data and points.min.
If min.A and min.B do not exist they are not created.
READMIN therefore provides an alternative way to start an initial
path calculation by providing minima, so long as min.A and min.B
are created by hand.
It also provides a way to add additional minima to an existing database, since
new entries should be appended. There is a check to make sure that the minima
added are different from all those currently known.
- READRATES nstates: will read in a rate matrix in the NGT procedure with nstates.
min.A and min.B are still required.
Element row i, column j should be the rate constant
ki←j
with minus the sum of rates out of minimum j in the diagonal entry j, j.
- READZSYM file: reads element symbols from file ZSYM in free format.
Having the correct symbols is required for XTB jobs unless there is only one atom type.
- NEWREAXFF: specifies the use of the REAXFF force field. The following additional files are needed:
fort.3 or start, which contain the starting geometry, fort.4, which contains
a specific force field parametrization, control, which contains additional force field settings.
We recommend using ReaxFF with a start file for the coordinates, in which case fort.3 will only
be used to read atomic symbols and number of atoms in the system. The format of fort.3 is BGF and is format-sensitive.
- REGROUP x: regroup the A and B sets using a disconnectivity graph
analysis [#!beckerk97!#,#!walesmw98!#,#!Wales03!#] at energy x.
Any intervening minima that share a superbasin with an A or B minimum are reclassified
to belong to the A and B sets, respectively.[#!TrygubenkoW06!#]
The number of minima and transition states does not change: only the
classification of minima as A or B is affected,
which will change any rate analysis that follows.
The corresponding
minima are printed in files min.A.regrouped and min.B.regrouped.
- REGROUPFREE x: the database will be regrouped by combining free
energy minima where forward and backward free energy barriers are both
less than x. Regrouping is performed iteratively until no further free
energy minima merge.
In this scheme, groups that contain product and reactant minima
(A and B sets) are not allowed to merge.
The regrouping continues even if product and reactant groups
could merge according to the barrier threshold: such mergers are simply forbidden.
Don't forget to assign an appropriate value for the Planck constant using the
PLANCK keyword.
If one of the DIJKSTRA, SDGT, SGT, or DGT keywords is
present the corresponding analysis will be performed using the new groups
and free energies.
- REGROUPFREEAB x: the database will be regrouped as for
REGROUPFREE above, but only the A and B groups are changed.
- REGROUPPE x: the database will be regrouped based upon
a superbasin analysis at potential energy x.
Minima will be reassigned as A and B-type if they are in a superbasin
with an A or B minimum for this threshold.
New files min.data.regrouped, ts.data.regrouped, min.A.regrouped
and min.B.regrouped will be created.
Don't forget to assign an appropriate value for the Planck constant using the
PLANCK keyword. See also REGROUPRATE.
REGROUPPE and REGROUPRATE are different from REGROUP
in that they actually change the internal database structure into free energy
minima and groups of transition states linking them.
In this scheme, groups that contain product and reactant minima
(A and B sets) are not allowed to merge.
The regrouping continues even if product and reactant groups
could merge according to the barrier threshold: such mergers are simply forbidden.
If one of the DIJKSTRA, SDGT, SGT, or DGT keywords is
present the corresponding analysis will be performed using the new groups
and free energies.
- REGROUPPERSIST thresh :
regrouping analogous to REGROUPFREE except that minima in groups defined
by the min.include file (which contains some of the most persistent minima, see
PERSIST) are also not allowed to merge, like the A and B
sets (unless ALLOWAB is set). The PERSIST keyword must also be present
(though the code could easily be changed to use any pre-existing min.include file...).
- REGROUPRATE x: the database will be regrouped based upon harmonic transition
state theory rate constants. Minima are in the same group if they can interconvert
via a path that involves only rate constants greater than x.
Minima will be reassigned as A and B-type if they are in a group
with an A or B minimum for this threshold.
New files min.data.regrouped, ts.data.regrouped, min.A.regrouped
and min.B.regrouped will be created.
Don't forget to assign an appropriate value for the Planck constant using the
PLANCK keyword. See also REGROUPPE.
REGROUPPE and REGROUPRATE are different from REGROUP
in that they actually change the internal database structure into free energy
minima and groups of transition states linking them.
In this scheme, groups that contain product and reactant minima
(A and B sets) are not allowed to merge.
The regrouping continues even if product and reactant groups
could merge according to the barrier threshold: such mergers are simply forbidden.
If one of the DIJKSTRA, SDGT, SGT, or DGT keywords is
present the corresponding analysis will be performed using the new groups
and free energies.
- REMOVESP: the minima specified in files min.remove and
ts.remove are removed and new files min.data.removed,
ts.data.removed, min.A.removed, min.B.removed,
points.min.removed and points.ts.removed
are created with a consistent numbering scheme for the remaining stationary
points. The first lines of min.remove and
ts.remove must contain a single integer, which is the number of
stationary points listed for removal on the remaining lines.
- REMOVEUNCONNECTED set : the database is rewritten to files
min.data.removed, points.min.removed, etc.
Minima disconnected from set, where set is A B or AB,
are removed, along with all transition states that connect them.
Minima with NCONNMIN connections or fewer are also removed (this
condition is applied recursively). The default for set is AB, which will remove
minima that have no connection to any member of the A or B sets.
- RETAINSP: minima not specified in file min.retain
are removed and new files min.data.retained,
ts.data.retained, min.A.retained, min.B.retained,
points.min.retained and points.ts.retained
are created with a consistent numbering scheme for the remaining stationary points.
The minima to be retained must appear in min.retain, one per line.
Transition states are only retained if they link two minima specified in the
min.retain file. Note, however, that at least one member each of
min.A and min.B must be included in min.retained. Otherwise,
ts.data.retained and points.ts.retained are not created.
Degenerate rearrangements do not affect the kinetics, so they are not retained.
- RETAINONLYSP: same as RETAINSP but
only the specified transition states in ts.retain are kept, so long as their minima are retained.
These transition states must be listed one per line in ts.retain.
We must also retain transition states that link the same minima as any specified in
ts.retain.
This step is essential for consistency with KDISTINCTPATHS
and KSHORTESTPATHS because connections between minima are indexed by only one of the possible
transition states,
namely the last one in the ts.data file. The branching probabilities include all contributions.
Unlike RETAINSP additional transition states linking retained minima are not included.
Degenerate rearrangements do not affect the kinetics, so they are not retained.
- REVERSEGEDIFFTOL: Reverses the order of comparison between two
structures (minima and transition states). First, their Cartesian distance will
be calculated and compared. If the distance is larger than GEOMDIFFTOL, the
structures are different. If the distance is less than GEOMDIFFTOL, the
energies of the two structures will be compared. If their energies differ by
more than EDIFFTOL, the structures are considered different, or else identical.
Typically, this leads to a slower calculation since the energies are already
present, while the distance comparison requires additional calculation.
Nevertheless, this is useful in cases where the energy landscape is noisy and
has a very rough topography (i.e. structurally very similar structures), which
is sometimes the case for ReaxFF force fields.
- REWEIGHT nrwbins nrwreactant energyfile:
conditional probabilities of reactant minima are obtained from the quench
probabilities calculated using the energies in file energyfile.
These energies could come from systematic quenching from an MD trajectory at
the same temperature, for example.
nrwbins is the number of bins to use in representing the quench
probability distribution and nrwreactant is the number of bins
to use in representing the minima in the reactant set.
This is a somewhat experimental option. Consult DJW before using!
- RELATIVEE: subtract the potential energy of the global minimum
from all the minima and transition states, as well as the threshold for
transition state neglect. This shift of the energy origin may
help to avoid some numerical underflow or overflow issues in rate
and thermodynamics calculations, especially at
low temperature.
It can only be used in post-processing, since we shift only the potential energy
values that are read in initially.
- RENORMREACTANT: in NGT calculations, distribute the equilibrium probability only
over connected reactants. This is not the default. It is needed for quantitative comparison
with rates obtained using eigendecomposition with WAITPDF, WAITPDFGEN, WAITPDFRANGE or WAITPDFSYM, where we
want to remove unconnected reactants to avoid zero eigenvalues.
- RFMULTI tau Tlow Thigh Tinc steps pfshift:
run the REGROUPFREE analysis as a function of temperature for a fixed
regrouping threshold. The threshold is determined from the observation time scale, tau
and the temperature range is Tlow to Thigh in steps
steps.
A fixed shift for the natural log of the partition functions can be specified
by the parameter pfshift, which may be needed to prevent underflow or overflow.
- RIGIDBODIES n: specifies the number of rigid bodies; only one
of NATOMS and RIGIDBODIES should be present. NB: this is not part of the GENRIGID framework.
- RIGIDINIT n: specifies the use of the generalised rigid body framework. You must also specify
the number of degrees of freedom in the rigidified system, n, so that PATHSAMPLE knows how many
eigenvalues to expect in OPTIM output.
- RUNLOCAL: for slurm batch jobs this keyword tells PATHSAMPLE to run the OPTIM
jobs in subdirectories of the submit directory, rather than copying all the files to node scratch space.
This setup works for CSD3 lustre file systems, but should not be used for the group clusters with nfs
local file systems.
- SAMEMIN: the minima specified in samemin as pairs, one pair per line, are
set to be the same in a new database. The algorithm sets the higher index minimum to be the same as
the lower index minimum specified, which may itself be equivalent to a lower index minimum, and so on.
A minimum is not allowed to be equivalent to two different lower index minima.
A lower index minimum is allowed to be equivalent to two different higher index minima.
PATHSAMPLE will write a new file ts.dosamemin with the replacement minima specified in the
transition state connections. This file is just for reference with the original minima numbers.
A new database is written using the REMOVESP procedure having written a min.remove file,
removing all the higher index equaivalent minima, and a ts.remove file with no entries.
The entries in samemin can be in any order.
- SBATCH: this option will submit OPTIM jobs using the sbatch command,
rather than running them as child processes with FORK or EXEC. A template sbatch script
called sbatch.template is required in the working directory. The job command corresponding
to the usual submit script is added to the end of the template. PATHSAMPLE will
not use WAIT, but simply cycle through the total number of OPTIM jobs corresponding
to the CYCLES and CPUS keywords.
Running PATHSAMPLE again with CHECKPATHINFO should enable runs to continue, picking
up the path.info files that have been generated. Queued OPTIM jobs could be
killed and replaced or new swarms added to the slurm queue.
This keyword was created to provide a way to use available GPU resources when the number of
cards available per node can vary. It could also provide a way to get around wall clock
limits on a parent PATHSAMPLE job, although the maximum run time would apply to each
OPTIM job instead.
- SDGT DisconnectSources AltPbb Rescale Normalise:
graph transformation rate calculation, which switches from sparse optimised to
dense optimised algorithms when the average number of connections
exceeds a certain threshold [#!TrygubenkoW06!#].
The four arguments are all logicals, so an example input line might look
SDGT T T F F. If true (the default) DisconnectSources specifies that
a full transformation should be performed for each source, disconnecting the
other sources. The resulting rate constants correspond to the KMC result and
kKMC [#!Wales06!#].
If AltPbb is true (default is false) then additional work is done to
maintain precision, which roughly doubles the execution time, but may be needed at low
temperature.
If Rescale is true (default false) an alternative strategy is used
to try and prevent error propagation.
Setting Normalise true (default false) instructs GT2input.f90 to check the normalisation
of branch probabilities. This should not be necessary.
- SEED n: random number seed.
- SGT DisconnectSources AltPbb Rescale Normalise:
sparse optimised graph transformation rate calculation [#!TrygubenkoW06!#].
The four arguments are all logicals, so an example input line might look
SGT T T F F. If true (the default) DisconnectSources specifies that
a full transformation should be performed for each source, disconnecting the
other sources. The resulting rate constants correspond to the KMC result and
kKMC [#!Wales06!#].
If AltPbb is true (default is false) then additional work is done to
maintain precision, which roughly doubles the execution time, but may be needed at low
temperature.
If Rescale is true (default false) an alternative strategy is used
to try and prevent error propagation.
Setting Normalise true (default false) instructs GT2input.f90 to check the normalisation
of branch probabilities. This should not be necessary.
- SHANNON tmin tmax tinc einc npeq:
calculate frustration measures for the landscape as a function of temperature,
starting from tmin (default 1), ending at tmax (default 2)
at intervals tinc (default 0.1).
einc is the energy interval for superbasin construction, used to estimate the
barriers.
npeq (default 100) is the number of minima to include in maintaining an ordered list of the
highest occupation probabilities.
These probabilities are printed, and they are used in the more expensive rate-based measures
that are computed for SHANNONR.
- SHANNONR tmin tmax tinc einc npeq:
calculate frustration measures for the landscape as a function of temperature,
starting from tmin (default 1), ending at tmax (default 2)
at intervals tinc (default 0.1).
As for SHANNON, but the more expensive rate-based metrics are calculated.
einc is the energy interval for superbasin construction, used to estimate the
barriers.
npeq (default 100) is the number of minima to include in maintaining an ordered list of the
highest occupation probabilities.
- SHIFTTS shift: the transition states listed in file shiftts have the value shift
added to their energies and a new file ts.data.shift is written containing the shifted values.
May be useful for DIJPATHPAIRS runs where transition state energies are shifted after the corresponding
minima have been tried. See also DIJPATHPAIRS and DNEBPATH.
- SHORTCUT minsep string maxsep: try connecting minima that are further apart than minsep
steps on the best path, as calculated using Dijkstra's algorithm.
In the Dijkstra analysis minima with zero connections are removed.
This value can be changed using a DIJKSTRA n line in pathdata.
For the string value MAXSEP a third parameter maxsep is needed, corresponding to the maximum
number of steps between minima for connection attempts. For very long paths a MAXSEP value
may be needed to prevent the distance calculation between pairs of minima from becoming a bottleneck.
The default is to consider all pairs, but this choice is probably not appropriate for very long paths.
If the option argument string is set to BARRIER or RATE
or MINMAX then
the strategy is different.
We now look for pairs of minima on either side of
the highest barriers or minima linked by the smallest
rate constant on the best path, sorted according to the barrier height or the rate.
minsep is then used to define the largest number of steps away from
the transition states for connection attempts.
If we run out of candidates for the fastest path then the highest energy transition state on
that path is blocked and a new fastest path is calculated until more candidates are obtained.
For MINMAX the minsep parameter is ignored and the pairs are chosen
as the lowest minima on either side of the barrier, i.e. the lowest minimum in the
discrete path before an uphill step as we move away from the transition state
in question. For DNEBPATH databases the lowest minima
could be the ones directly connected to the transition state in question, and these
are allowed to be chosen if the DNEBPATH keyword is also set.
- SKIPPAIRS : do not attempt to connect a pair of minima if they
are already connected. This is used only with USEPAIRS. Pairs which are
rejected on these grounds DO count towards the total number of connections
provided by the CYCLES keyword, however the debug printing doesn't recognise
that at the present time.
- SKIPTS : skip transition states specified in file ts.skip.
This keyword is intended for post-processing kinetic analysis only, so the number
of cycles must be zero. The points files should not be used in such calculations.
- SLEEPTIME x : set the time used in the system sleep calls
between submission of OPTIM jobs. The default value is 1 s. A nonzero
value is needed for small systems on distributed memory machines to prevent
us from running out of ports. A value of zero on a shared memory system can
speed up execution significantly for small systems.
- SLURM : OPTIM jobs are submitted to nodes using srun when the
queueing system is SLURM. This is intended to replace the use of keywords SSH
and PBS on clusters using SLURM. Job submission using ssh causes problems with
CUDAOPTIM jobs on pat, so keyword SLURM must be used alongside keyword CUDA.
- SSH : OPTIM jobs are submitted to nodes using ssh rather than rsh (default)
- ST : calls a finite system of Stockmayer particles.
- STARTFROMPATH afile n1 n2 : reads an initial path from file afile.
The files min.A and min.B are created: min.A contains the entries
1 and n1 and min.B contains the entries 1 and n2.
If min.A already exists then the run will terminate with a warning message
without overwriting it.
- SYSTEM a: the atom type label to be written to odata files for OPTIM,
thus specifying the potential that OPTIM will use.
- TAG n x: atom number n in the system is `tagged' by giving it
an artificial mass x.
- TEMPERATURE x: specifies that the canonical ensemble at reduced temperature x
is to be used to calculate rate constants. Mutually exclusive with ENERGY.
- TRAP: specifies a trapped ion potential. This keyword is needed to set the
number of zero eigenvalues. See the corresponding keyword in OPTIM for more information.
- TSTHRESH x: specifies that transition states above energy x should
not be included in the database. Probably most useful with CHARMM, where genuine
transition states with silly energies can certainly occur and give rise to underflow.
- TWOD: the system is two-dimensional.
- UNRES: specifies the UNRES potential.[#!liwoopwrs97!#]
- UNTRAP einc thresh edeltamin elowbar ehighbar: try connecting minima to eliminate traps.
Connections are attempted between all minima and any of the minima in the
product set based on the largest barriers in the product direction, as estimated
from disconnectivity analysis at energy intervals of einc, starting from the
global minimum.
The product basin is defined as the set at the highest energy for which the
minimum in question is not a member.
Candidate pairs of minima are sorted based upon the ratio of the
potential energy barrier to the potential energy difference between the minimum
in question and the lowest product minimum.
Hence this scheme is analogous to the FREEPAIRS procedure, but
uses individual minima and potential energy, instead of free energy.
The original product minimum may be replaced by a minimum from
the same superbasin that is closer to the target minimum, so long as it
lies within an energy thresh above the minimum to be untrapped.
Minima with zero connections are not considered;
this value can be changed using a DIJKSTRA n line in pathdata, even
though a Dijkstra analysis is not used.
See also the PAIRLIST keyword.
The UNTRAP keyword may provide the best way to remove artificial
frustration from potential energy disconnectivity graphs.
It may be helpful to choose a larger value of einc than would be
used for the superbasin analysis in visualising the disconnectivity graph.
This will encourage connection attempts with minima of lower energy in the
expanded product superbasin.
The optional arguments edeltamin and elowbar are useful for databases
without a well-defined global minimum. If the potential energy difference between the minimum
in question and the lowest product minimum is less than edeltamin, the value of
edeltamin is substituted for this difference. This substitution prevents
connection attempts to minima connected by low barriers but with a very small
energy difference to the lowest product minimum. If the barrier from the lowest product minimum to the minimum in question is less than elowbar, a value of TINY(1.0D0) is given to the barrier instead. This substitution prevents connection attempts to minima connected
by low barriers.
The fifth optional argument ehighbar aids with the selection of minima for
untrapping if you wish to target particular regions. Minima with a barrier height (in the direction
of the lowest-energy product minimum) larger than EHIGHBAR will not be selected.
Sometimes there are large groups of minima separated by small barriers that form a trap. We only need to find a lower-energy path for one of these minima and untrapping all of them may be a wasted effort. Running disconnectionDPS with the keyword BASINT followed by a particular energy gives the results of a basin analysis at this energy in a file `basins'. If this file is renamed as `noduplicatebasins' and placed in the directory where the PATHSAMPLE job is running, untrap will only pick one minimum from this group for untrapping.
- UNTRAPMETRIC nitems nmatrix baildist: to be used with UNTRAP. nitems is the number of metrics to use (1,2 or 3), nmatrix is the number of closest minima for which distances are calculated (default 2000) and baildist is a distance limit, if any distance is found to be below this limit, no further pairs are tried (default TINY(1.0D0)).
To use this keyword a `metric' file is required. The `metric' file takes the following format, with one line entry for each minimum in the database:
metric1 nmin [metric2] [metric3], metric1 takes priority over metric 2 etc...
You have to construct this file manually with your own choice of metric(s).
You also need a `metric.ordered' file - this speeds up the finding of pairs.
In /SCRIPTS/PATHSAMPLE/ there is a program to convert the metric file and produce a metric.ordered file, `getmetricordered.f90'.
However, you won't normally want all minima in the `metric.ordered' file. Ideally, this is a list of minima in the main basin to which connection attempts should be made, usually the main basin can be defined by an high energy cutoff.
Running disconnectionDPS with the keyword BASINT followed by a particular energy gives the results of a basin analysis at this energy in a file `basins'.
`getmetricordered.f90' will produce a `metric.ordered' file from `metric' and `basins', if you set the required input arguments.
The metric and metric.ordered files must be constructed manually and are currently not updated during a run when minima are added to the database.
Different metrics are required for different systems.
- USEEXEC value: value shall be either TRUE or FALSE (default TRUE).
If TRUE, after PATHSAMPLE has forked, the child PATHSAMPLE will use
an exec call to replace itself with the OPTIM job, rather than running the OPTIM
job as a subprocess. This should save system resources. This is only implemented
with SLURM, so the keyword has no effect if SLURM is not used. This is now the default
behaviour with SLURM, so specifying FALSE here is the only way to get the child
PATHSAMPLE to run OPTIM as a subprocess instead.
- USEPAIRS Epathfile: pairs of minima are chosen for connection attempts based
on the order of local minima specified in file Epathfile, which must have the format
of a PATHSAMPLE Epath output file 9as generated by DIJKSTRA, etc.
This keyword is intended to follow a DUMMYTS run, where the stationary point
database has been refined using BHINTERP or BISECT runs in OPTIM.
The dummy ts.data and pairs.data files must be renamed or removed
before the USEPAIRS run.
The pairs of minima are first chosen to be the adjacent structures from the list, then
second-neighbours, until all possible pairs have been tried.
This keyword can also be used to specify a list of pairs to attempt to connect. See the MAKEPAIRS keyword to help assemble Epathfile in this case.
- WAITPDF and WAITPDFSYM: perform an eigendecomposition and waiting time analysis using
absorbing product boundary conditions.
Each temperature, waiting time moments, and forward and backward rate constants are dumped to
file wait.rates.
The waiting time probability distribution functions are dumped to
waitpdfBA and waitpdfAB.
The waiting time accumulated as a sum over eigenmodes is dumped in
tauAB and tauBA.
For WAITPDFSYM the rate matrix is symmetrised before diagonalisation.
- WAITPDFRANGE il iu: perform an eigendecomposition and waiting time analysis using
absorbing product boundary conditions as for WAITPDFSYM.
Additional checks and features are used.
The upper and lower eigenvalues can be specified explicitly with il and iu,
which default to one and the maximum range.
The eigenvalues are ordered smallest to largest, so the slowest relaxations are the largest
because the eigenvalues should be negative.
Eigensolutions are checked and rejected if the eigenvalue is negative, if the sum of amplitudes
exceeds one, or if the mean first passage time (MFPT) deviates for the NGT value by more that a factor
of e. The last of these checks requires NGT to be run for the same temperatures and for the same source
minimum, which requires lines in pathdata such as:
WAITPDFRANGE
WAITPDFTEMPS 0.6 0.7
RATESCYCLE 0.6 0.7
NGT 0 T 1.1D0 1 100.1D0
WAITSTARTMIN 604
NGTSTARTMIN 604
If the MFPT is available from NGT, and one or more eigenvalues for slow relaxations are rejected,
an additional term is added to the first passage time distributions for AB and BA.
The amplitude is one minus the sum of amplitudes for accepted eigenvalues, and the missing eigenvalue
is chosen to give the same MFPT as the NGT value.
Changing the upper limit iu enables the NGT MFPT construction for the last peak of the first
passage time distribution to be tested.
- WAITPDFGEN: solve the full master equation using eigendecomposition with rates out of sink states
set to zero. The sink states are the A minima and the sources are the B minima. The initial probability is
defined by the relative equilibrium values for the souce minima.
In this implementation the occupation probabilities for all states are available as a function of time.
The first passage time distribution should be the same as for WAITPDF and WAITPDFSYM.
With this keyword the final relative probabilities in the different sinks are available, and depend
on the initial conditions (no unique equilibrium because there are additional zero eiegenvalues in the
rate matrix). We also dump the mean energy as a function of time in mean_energy and
mean_energy2. The values in the second file are the conditional average restricted to the non-sink states.
This average can track the values for distinct traps on the landscape that appear in the FPT distribution.
- WAITPDFTEMPS t1 t2 ... tn : perform the eigendecomposition and
waiting time analysis for temperatures t1, t2, etc.
Each temperature, waiting time moments, and forward and backward rate constants are dumped to
file wait.rates.
The waiting time probability distribution functions are dumped to
waitpdfBA and waitpdfAB.
The waiting time accumulated as a sum over eigenmodes is dumped in
tauAB and tauBA.
Any number of temperatures can be specified, so long as continuation
lines are used if the line length exceeds 80 characters.
See also WAITPDF, WAITPDFSYM, and ARNOLD.
One of WAITPDF or WAITPDFSYM must be specified.
- XTB: Specify that XTB is used. PATHSAMPLE will write start.* files for OPTIM rather than appending the coordinates to the odata files. A xtb.symbols file must be present containing the element symbols for all atoms (one per line).
 and
and
 , respectively.
, respectively.
![\includegraphics[width=0.5\textwidth]{PHE.eps}](img4.png)
 ). The usual criteria for transition
states and the connectivity of minima also apply.
The file min.include has a format suitable for reading via the
PICK and IDMINFILE keywords in disconnectionDPS. The format of the min.persist* files
is: rank (ordered by persistence), energy, energy+barrier, barrier(=persistence), index of the minimum,
deviation from the diagonal. See also PERSISTEXACT
). The usual criteria for transition
states and the connectivity of minima also apply.
The file min.include has a format suitable for reading via the
PICK and IDMINFILE keywords in disconnectionDPS. The format of the min.persist* files
is: rank (ordered by persistence), energy, energy+barrier, barrier(=persistence), index of the minimum,
deviation from the diagonal. See also PERSISTEXACT
![[*]](crossref.png) for more details.
for more details.